Structure of the E. coli protein-conducting channel bound to a translating ribosome - PubMed (original) (raw)
Structure of the E. coli protein-conducting channel bound to a translating ribosome
Kakoli Mitra et al. Nature. 2005.
Abstract
Secreted and membrane proteins are translocated across or into cell membranes through a protein-conducting channel (PCC). Here we present a cryo-electron microscopy reconstruction of the Escherichia coli PCC, SecYEG, complexed with the ribosome and a nascent chain containing a signal anchor. This reconstruction shows a messenger RNA, three transfer RNAs, the nascent chain, and detailed features of both a translocating PCC and a second, non-translocating PCC bound to mRNA hairpins. The translocating PCC forms connections with ribosomal RNA hairpins on two sides and ribosomal proteins at the back, leaving a frontal opening. Normal mode-based flexible fitting of the archaeal SecYEbeta structure into the PCC electron microscopy densities favours a front-to-front arrangement of two SecYEG complexes in the PCC, and supports channel formation by the opening of two linked SecY halves during polypeptide translocation. On the basis of our observation in the translocating PCC of two segregated pores with different degrees of access to bulk lipid, we propose a model for co-translational protein translocation.
Figures
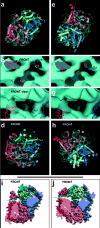
Figure 1
General features of the cryo-EM reconstruction of E. coli RNC-SecYEG complex. Cryo-EM densities are shown for the small (30S, yellow) and large (50S, sky-blue) subunits, and the A-, P- and E-site tRNAs (magenta, green, and orange). Isolated densities for the mRNA (cyan), the nascent chain in the ribosomal polypeptide exit tunnel (gold), the protein-conducting channel (PCC) at the polypeptide exit site (translocating PCC, blue) and the PCC at the 5′ mRNA channel exit (non-translocating PCC, red) are also shown. a, General overview of the RNC-SecYEG complex. b,c, Closeup of the translocating PCC, shown in the front view (b) with the nascent chain visible through the large frontal opening between the ribosome and the PCC (gold arrow). The line of view is perpendicular to the plane of the membrane (c). d, e, Closeups of the non-translocating PCC.
Figure 2
Normal mode-based flexible fitting (NMFF) of the SecYEG complex into cryo-EM density. a,d, Model after NMFF of a front-to-front arrangement of two SecYEG_Ec_ heterotrimers into the non-translocating (a) and translocating (b) PCC density. b,c, Placement of the non-translocating front-to-front model into the translocating PCC density, such that the ribosome-binding SecY cytoplasmic loops are placed within connection densities. The three connection regions (C1–C3) are shown in cross-section (line of view perpendicular to the membrane plane) (b) and in the front view (c), with the frontal opening visible between C1 and C2. EM density is shown in cyan as a semi-transparent surface. e–h, Parallel analysis of the back-to-back SecYEG_Ec_ model. i,j, van der Waals surface representations of the front-to-front models obtained by fitting to the non-translocating (i) and translocating PCC density (j) (C-terminal SecY halves transparent). The green and yellow arrows indicate the change in the heterotrimer interface at the front, and linked SecY half opening, respectively. In the atomic models one heterotrimer is colored in shades of blue-green, the other in shades of red. The view is within the plane of the membrane, with the ribosomal side behind the plane. In a,d,e,h, atomic models, with TMHs depicted as rods, are shown docked into the experimental EM density (cyan mesh).
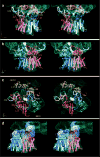
Figure 3
Stereo views of RNA and protein elements in the ribosome-PCC junction. Real-space refined models of E. coli ribosomal proteins are rendered as ribbons and ribosomal RNA regions interacting with the PCC as thick, light-grey backbone rattlers. The PCC is colored and rendered as in Fig. 2d with the cryo-EM density in cyan mesh. ac, The ribosome-PCC junction at the polypeptide exit site (nascent chain density semi-transparent yellow surface) is shown in the front (a) and back (b) views. c, Ribosomal elements near the polypeptide exit site and parts of the PCC in the three connection regions are illustrated. The line of view is perpendicular to the membrane plane. Connection regions between ribosome and PCC are circled in orange and labeled. d, The non-translocating PCC interacts via its CFADs with hairpins in the mRNA (semi-transparent purple surface with non-interacting mRNA rattler regions in yellow).
Figure 4
The path of the nascent chain through the ribosome and PCC. a, Fitting of a front-to-front SecYEG_Ec_ model into the translocating PCC EM density leaves prominent regions of density unaccounted for (green and yellow asterisks). The PCC is viewed within the plane of the membrane, with the ribosome behind the plane, and colored/rendered as before with TMHs numbered. b, The nascent chain (yellow rattler) was fitted into the isolated polypeptide density inside the ribosome. The front view is shown. Schematized versions of the PCC at the polypeptide exit site of the ribosome in views corresponding to a (c) and b (d). The PCC and ribosomal elements are colored as before, with the nascent chain in green (TMH signal anchor) and yellow. See text for discussion.
Comment in
- Cell biology: two pores better than one?
Driessen AJ. Driessen AJ. Nature. 2005 Nov 17;438(7066):299-300. doi: 10.1038/438299a. Nature. 2005. PMID: 16292298 No abstract available.
Similar articles
- Structure of the SecY channel during initiation of protein translocation.
Park E, Ménétret JF, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW. Park E, et al. Nature. 2014 Feb 6;506(7486):102-6. doi: 10.1038/nature12720. Epub 2013 Oct 23. Nature. 2014. PMID: 24153188 Free PMC article. - Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Frauenfeld J, et al. Nat Struct Mol Biol. 2011 May;18(5):614-21. doi: 10.1038/nsmb.2026. Epub 2011 Apr 17. Nat Struct Mol Biol. 2011. PMID: 21499241 Free PMC article. - Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Gogala M, Becker T, Beatrix B, Armache JP, Barrio-Garcia C, Berninghausen O, Beckmann R. Gogala M, et al. Nature. 2014 Feb 6;506(7486):107-10. doi: 10.1038/nature12950. Nature. 2014. PMID: 24499919 - The structure of the bacterial protein translocation complex SecYEG.
Collinson I. Collinson I. Biochem Soc Trans. 2005 Dec;33(Pt 6):1225-30. doi: 10.1042/BST0331225. Biochem Soc Trans. 2005. PMID: 16246086 Review. - The machinery of membrane protein assembly.
White SH, von Heijne G. White SH, et al. Curr Opin Struct Biol. 2004 Aug;14(4):397-404. doi: 10.1016/j.sbi.2004.07.003. Curr Opin Struct Biol. 2004. PMID: 15313232 Review.
Cited by
- Dynamic structure of the translocon SecYEG in membrane: direct single molecule observations.
Sanganna Gari RR, Frey NC, Mao C, Randall LL, King GM. Sanganna Gari RR, et al. J Biol Chem. 2013 Jun 7;288(23):16848-16854. doi: 10.1074/jbc.M113.471870. Epub 2013 Apr 22. J Biol Chem. 2013. PMID: 23609442 Free PMC article. - Molecular dynamics studies of the archaeal translocon.
Gumbart J, Schulten K. Gumbart J, et al. Biophys J. 2006 Apr 1;90(7):2356-67. doi: 10.1529/biophysj.105.075291. Epub 2006 Jan 13. Biophys J. 2006. PMID: 16415058 Free PMC article. - Electrophysiological studies in Xenopus oocytes for the opening of Escherichia coli SecA-dependent protein-conducting channels.
Lin BR, Gierasch LM, Jiang C, Tai PC. Lin BR, et al. J Membr Biol. 2006;214(2):103-13. doi: 10.1007/s00232-006-0079-1. Epub 2007 May 25. J Membr Biol. 2006. PMID: 17530158 Free PMC article. - Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome.
Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Becker T, et al. Science. 2009 Dec 4;326(5958):1369-73. doi: 10.1126/science.1178535. Epub 2009 Oct 29. Science. 2009. PMID: 19933108 Free PMC article. - Environmental transition of signal-anchor sequences during membrane insertion via the endoplasmic reticulum translocon.
Kida Y, Kume C, Hirano M, Sakaguchi M. Kida Y, et al. Mol Biol Cell. 2010 Feb 1;21(3):418-29. doi: 10.1091/mbc.e09-08-0738. Epub 2009 Dec 2. Mol Biol Cell. 2010. PMID: 19955210 Free PMC article.
References
- Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. - PubMed
- Wickner W, Driessen AJM, Hartl FU. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem. 1991;60:101–124. - PubMed
- Brundage L, et al. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. - PubMed
- Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–30. - PubMed
- Gilmore R, Blobel G. Translocation of secretory proteins across the microsomal membrane occurs through an environment accessible to aqueous perturbants. Cell. 1985;42:497–505. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR001219/RR/NCRR NIH HHS/United States
- P41 RR012255/RR/NCRR NIH HHS/United States
- R01 GM055440/GM/NIGMS NIH HHS/United States
- R37 GM029169/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases