Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set - PubMed (original) (raw)
. 2006 Jan;188(2):759-72.
doi: 10.1128/JB.188.2.759-772.2006.
Alifiya S Motiwala, Magali Cavatore, Weihong Qi, Manzour Hernando Hazbón, Miriam Bobadilla del Valle, Janet Fyfe, Lourdes García-García, Nalin Rastogi, Christophe Sola, Thierry Zozio, Marta Inírida Guerrero, Clara Inés León, Jonathan Crabtree, Sam Angiuoli, Kathleen D Eisenach, Riza Durmaz, Moses L Joloba, Adrian Rendón, José Sifuentes-Osornio, Alfredo Ponce de León, M Donald Cave, Robert Fleischmann, Thomas S Whittam, David Alland
Affiliations
- PMID: 16385065
- PMCID: PMC1347298
- DOI: 10.1128/JB.188.2.759-772.2006
Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set
Ingrid Filliol et al. J Bacteriol. 2006 Jan.
Erratum in
- J Bacteriol. 2006 Apr;188(8):3162-3
Abstract
We analyzed a global collection of Mycobacterium tuberculosis strains using 212 single nucleotide polymorphism (SNP) markers. SNP nucleotide diversity was high (average across all SNPs, 0.19), and 96% of the SNP locus pairs were in complete linkage disequilibrium. Cluster analyses identified six deeply branching, phylogenetically distinct SNP cluster groups (SCGs) and five subgroups. The SCGs were strongly associated with the geographical origin of the M. tuberculosis samples and the birthplace of the human hosts. The most ancestral cluster (SCG-1) predominated in patients from the Indian subcontinent, while SCG-1 and another ancestral cluster (SCG-2) predominated in patients from East Asia, suggesting that M. tuberculosis first arose in the Indian subcontinent and spread worldwide through East Asia. Restricted SCG diversity and the prevalence of less ancestral SCGs in indigenous populations in Uganda and Mexico suggested a more recent introduction of M. tuberculosis into these regions. The East African Indian and Beijing spoligotypes were concordant with SCG-1 and SCG-2, respectively; X and Central Asian spoligotypes were also associated with one SCG or subgroup combination. Other clades had less consistent associations with SCGs. Mycobacterial interspersed repetitive unit (MIRU) analysis provided less robust phylogenetic information, and only 6 of the 12 MIRU microsatellite loci were highly differentiated between SCGs as measured by GST. Finally, an algorithm was devised to identify two minimal sets of either 45 or 6 SNPs that could be used in future investigations to enable global collaborations for studies on evolution, strain differentiation, and biological differences of M. tuberculosis.
Figures
FIG. 1.
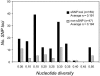
Nucleotide diversity among SNPs identified through intergenomic comparisons. The distribution of nucleotide diversity for 159 sSNPs and 47 non-sSNPs among 219 strains of M. tuberculosis and M. bovis for which complete SNP data were available.
FIG. 2.
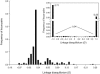
Linkage disequilibrium among study loci. The distribution of the linkage disequilibrium coefficient (D) for 12,246 pairwise comparisons of alleles at 159 sSNP loci. A total of 5,822 (47%) of these comparisons are significant by a chi-squared test, and 3,008 (52%) remained significant using a Bonferroni correction for multiple tests. The insert shows the distribution of the standardized coefficient of linkage disequilibrium (_D_′). Ninety-six percent of the locus pairs are in complete linkage disequilibrium (i.e., _D_′ = 1).
FIG. 3.
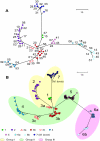
Phylogenetic trees of a global collection of M. tuberculosis isolates. A. Distance-based neighbor joining tree of M. tuberculosis and M. bovis based on 159 sSNPs resolved the 219 isolates into 56 sequence types (STs). Each ST is indicated by a dot color coded for membership in an SCG and a numerical value. The bootstrap values associated with SCG-1 and SCG-2 to SCG-7 are 84% and 99%, respectively. Reference strains M. tuberculosis 210, M. tuberculosis CDC1551, M. tuberculosis H37Rv, and M. bovis AF2122/97 are present in SCG-2, -4, -6b, and -7, respectively. The modeling-based clustering analysis of the same comprehensive set of the genotype data (159 sSNPs in 219 isolates) identified the same seven clusters. B. Model-based neighbor-joining tree based on a comprehensive data set of 212 SNPs resolved the 327 isolates into 182 STs and presented identical clusters as shown in panel A. Numbers designate each SCG; SC subgroups are indicated by colored symbols. The SNP lineages that belong to the three “major genetic” groups based on combination of two alleles at katG463 and gyrA95 are also highlighted. The scale bar indicates the number of SNP differences.
FIG. 4.
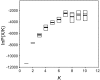
Inferring the number of clusters in an M. tuberculosis phylogeny. The number of clusters (K) were inferred for the genotype data of 219 M. tuberculosis and M. bovis strains on the basis of 159 sSNPs using the model-based clustering method implemented in STRUCTURE. Five independent simulations were run for K values ranging from 1 to 10. The top and bottom boundaries of each box indicate the 25th percentile and the 75th percentile, respectively. The thick black line and the thin black line within the box mark the median and the mean, respectively. The estimate of the posterior probability of K, P(X K), was the highest when K = 7.
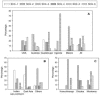
FIG. 5.
Distribution of the SCGs by country of origin. The proportion of M. tuberculosis isolates with a particular SCG is shown for various geographic regions. A. The country where the isolate was cultured. B. The place of birth of the human host from which the isolate was cultured for isolates obtained from Australia. C. The place of birth of the human host from which the isolate was cultured for isolates obtained from Mexico. “Indian subcontinent” includes M. tuberculosis isolates from India, Pakistan, and Afghanistan and seven additional M. tuberculosis isolates obtained independently from India. “East Asia” includes M. tuberculosis isolates from Vietnam, China, Philippines, Indonesia, Korea, Hong Kong, and Laos. “Others” includes M. tuberculosis isolates from Australia, Europe, and Africa. Huauchinango and Orizaba represent relatively closed populations, while Monterey represents a more urban population. Two Mexican isolates for which the origin of host was unknown were not included.
FIG. 6.
Distribution of the spoligotype clades on the SNP-based phylogeny. Each isolate is indicated by a dot, which is color coded according to the spoligotype clade assignment. CAS, Central Asian clade; EAI, East African-Indian clade; H, Haarlem clade; LAM, Latin American and Mediterranean clade; PINI, Mycobacterium pinnipedii clade; S, S clade; T, T clade; X, X clade.
FIG. 7.
Distribution of SCGs on the distance-based neighbor-joining tree representing the MIRU phylogeny. SCG assignment (SCG-1 through SCG-7) of each MIRU type is based on the same colors and symbols used in Fig. 3B. The location(s) of the SCG-3a isolates (red triangles) are not shown, because MIRU results were not available for this group.
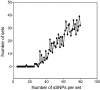
FIG. 8.
Determining the size of the minimal SNP set. Graph representing the number of sSNP sets that give the same cluster structure as that inferred from the comprehensive set of the 159 sSNPs versus the number of sSNP loci per set (ranging from 5 to 80). For each genotype length, sSNPs were generated by 100 random samples from the original source of 159 sSNPs. Cluster structure was inferred by the model-based method in STRUCTURE. In this simulation study, the minimum number of sSNP loci needed to resolve the same clusters was 16.
Similar articles
- Single nucleotide polymorphisms in Mycobacterium tuberculosis and the need for a curated database.
Stucki D, Gagneux S. Stucki D, et al. Tuberculosis (Edinb). 2013 Jan;93(1):30-9. doi: 10.1016/j.tube.2012.11.002. Epub 2012 Dec 21. Tuberculosis (Edinb). 2013. PMID: 23266261 Free PMC article. Review. - A snapshot of the predominant single nucleotide polymorphism cluster groups of Mycobacterium tuberculosis clinical isolates in Delhi, India.
Varma-Basil M, Narang A, Chakravorty S, Garima K, Gupta S, Kumar Sharma N, Giri A, Zozio T, Couvin D, Hanif M, Bhatnagar A, Menon B, Niemann S, Rastogi N, Alland D, Bose M. Varma-Basil M, et al. Tuberculosis (Edinb). 2016 Sep;100:72-81. doi: 10.1016/j.tube.2016.07.007. Epub 2016 Jul 25. Tuberculosis (Edinb). 2016. PMID: 27553413 - Genetic Diversity of Mycobacterium tuberculosis Isolates from Assam, India: Dominance of Beijing Family and Discovery of Two New Clades Related to CAS1_Delhi and EAI Family Based on Spoligotyping and MIRU-VNTR Typing.
Devi KR, Bhutia R, Bhowmick S, Mukherjee K, Mahanta J, Narain K. Devi KR, et al. PLoS One. 2015 Dec 23;10(12):e0145860. doi: 10.1371/journal.pone.0145860. eCollection 2015. PLoS One. 2015. PMID: 26701129 Free PMC article. - Can 15-locus mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis provide insight into the evolution of Mycobacterium tuberculosis?
Gibson A, Brown T, Baker L, Drobniewski F. Gibson A, et al. Appl Environ Microbiol. 2005 Dec;71(12):8207-13. doi: 10.1128/AEM.71.12.8207-8213.2005. Appl Environ Microbiol. 2005. PMID: 16332804 Free PMC article. - [New era in molecular epidemiology of tuberculosis in Japan].
Takashima T, Iwamoto T. Takashima T, et al. Kekkaku. 2006 Nov;81(11):693-707. Kekkaku. 2006. PMID: 17154049 Review. Japanese.
Cited by
- Clonality and micro-diversity of a nationwide spreading genotype of Mycobacterium tuberculosis in Japan.
Wada T, Iwamoto T, Tamaru A, Seto J, Ahiko T, Yamamoto K, Hase A, Maeda S, Yamamoto T. Wada T, et al. PLoS One. 2015 Mar 3;10(3):e0118495. doi: 10.1371/journal.pone.0118495. eCollection 2015. PLoS One. 2015. PMID: 25734518 Free PMC article. - Genetic diversity and molecular epidemiology of multidrug-resistant Mycobacterium tuberculosis in Minas Gerais State, Brazil.
Dantas NG, Suffys PN, Carvalho Wda S, Gomes HM, de Almeida IN, de Assis LJ, Augusto CJ, Gomgnimbou MK, Refregier G, Sola C, de Miranda SS. Dantas NG, et al. BMC Infect Dis. 2015 Aug 1;15:306. doi: 10.1186/s12879-015-1057-y. BMC Infect Dis. 2015. PMID: 26231661 Free PMC article. - Single nucleotide polymorphisms in Mycobacterium tuberculosis and the need for a curated database.
Stucki D, Gagneux S. Stucki D, et al. Tuberculosis (Edinb). 2013 Jan;93(1):30-9. doi: 10.1016/j.tube.2012.11.002. Epub 2012 Dec 21. Tuberculosis (Edinb). 2013. PMID: 23266261 Free PMC article. Review. - DNA polymorphisms in the pepA and PPE18 genes among clinical strains of Mycobacterium tuberculosis: implications for vaccine efficacy.
Hebert AM, Talarico S, Yang D, Durmaz R, Marrs CF, Zhang L, Foxman B, Yang Z. Hebert AM, et al. Infect Immun. 2007 Dec;75(12):5798-805. doi: 10.1128/IAI.00335-07. Epub 2007 Sep 24. Infect Immun. 2007. PMID: 17893137 Free PMC article. - The past and future of tuberculosis research.
Comas I, Gagneux S. Comas I, et al. PLoS Pathog. 2009 Oct;5(10):e1000600. doi: 10.1371/journal.ppat.1000600. Epub 2009 Oct 26. PLoS Pathog. 2009. PMID: 19855821 Free PMC article. Review.
References
- Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. - PubMed
- Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. - PMC - PubMed
- Barczak, A. K., P. Domenech, H. I. Boshoff, M. B. Reed, C. Manca, G. Kaplan, and C. E. Barry III. 2005. In vivo phenotypic dominance in mouse mixed infections with Mycobacterium tuberculosis clinical isolates. J. Infect. Dis. 192:600-606. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous