Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana - PubMed (original) (raw)
Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana
Jingdan Shen et al. Genetics. 2006 Feb.
Abstract
While the presence/absence polymorphism is commonly observed in disease resistance (R-) genes in Arabidopsis, only a few R-genes under the presence/absence polymorphism (R-P/A) have been investigated. To understand the mechanism of the molecular evolution of R-P/A, we investigated genetic variation of nine R-P/A in A. thaliana from worldwide populations. The number of possessed R-genes varied widely among accessions (two to nine, on average 4.3 +/- 1.6/accession). No pair of accessions shared the same haplotype, and no clear geographic differentiation was observed with respect to the pattern of presence/absence of the R-genes investigated. Presence allele frequencies also varied among loci (25-70%), and no linkage disequilibrium was detected among them. Although the LRR region in regular R-genes is known to be highly polymorphic and has a high Ka/Ks ratio in A. thaliana, nucleotide sequences of this region in the R-P/A showed a relatively low level of genetic variation (pi = 0.0002-0.016) and low Ka/Ks (0.03-0.70, <1). In contrast, the nucleotide diversities around the deletion junction of R-P/A were constantly high between presence and absence accessions for the R-genes (D(xy) = 0.031-0.103). Our results suggest that R-P/A loci evolved differently from other R-gene loci and that balancing selection plays an important role in molecular evolution of R-P/A.
Figures
Figure 1.
Schematic comparison of _R-_gene presence and absence alleles. Insertions with the _R_-gene are represented for At4g10780 (A), At5g05400 (B), At5g49140 (C), At5g18350 (D), Rpm1 (E), and Rps5 (F). (E and F are drawn on the basis of reports by H
enk
et al. 1999 and G
rant
et al. 1998.) In all absence alleles examined, the _R-_genes were replaced by 98–∼639-bp junk DNA of unknown origin.
Figure 2.
Grouping of A. thaliana accessions with respect to the presence/absence of the nine R-P/A. This tree was constructed by the neighbor-joining method using PAUP*4.0 (S
wofford
2000).
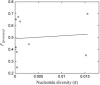
Figure 3.
Relationship between presence allele frequency (_F_[presence]) and nucleotide diversity of the LRR region in the nine R-P/A. Correlation coefficient was 0.093.
Figure 4.
Phylogenetic relationship of _R_-genes in the Col genome in A. thaliana. The tree was dissected from the one constructed by M
eyers
et al. (2003), using amino acid sequences of the NBS domain of _R_-genes by the neighbor-joining method. Nine R-P/A loci investigated in this study are indicated by boldface type. The Streptomyces sequence rooted the tree as the outgroup (M
eyers
et al. 2003).
Similar articles
- A genome-wide survey of R gene polymorphisms in Arabidopsis.
Bakker EG, Toomajian C, Kreitman M, Bergelson J. Bakker EG, et al. Plant Cell. 2006 Aug;18(8):1803-18. doi: 10.1105/tpc.106.042614. Epub 2006 Jun 23. Plant Cell. 2006. PMID: 16798885 Free PMC article. - Contrasting patterns of evolution between allelic groups at a single locus in Arabidopsis.
Ding J, Cheng H, Jin X, Araki H, Yang Y, Tian D. Ding J, et al. Genetica. 2007 Mar;129(3):235-42. doi: 10.1007/s10709-006-0002-9. Epub 2006 Aug 16. Genetica. 2007. PMID: 16912841 - Unusual signatures of highly adaptable R-loci in closely-related Arabidopsis species.
Wang J, Zhang L, Li J, Lawton-Rauh A, Tian D. Wang J, et al. Gene. 2011 Aug 15;482(1-2):24-33. doi: 10.1016/j.gene.2011.05.012. Epub 2011 Jun 1. Gene. 2011. PMID: 21664259 - Signature of balancing selection in Arabidopsis.
Tian D, Araki H, Stahl E, Bergelson J, Kreitman M. Tian D, et al. Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11525-30. doi: 10.1073/pnas.172203599. Epub 2002 Aug 9. Proc Natl Acad Sci U S A. 2002. PMID: 12172007 Free PMC article. - The arms race is ancient history in Arabidopsis, the wildflower.
Holub EB. Holub EB. Nat Rev Genet. 2001 Jul;2(7):516-27. doi: 10.1038/35080508. Nat Rev Genet. 2001. PMID: 11433358 Review.
Cited by
- Molecular evolution of the rice blast resistance gene Pi-ta in invasive weedy rice in the USA.
Lee S, Jia Y, Jia M, Gealy DR, Olsen KM, Caicedo AL. Lee S, et al. PLoS One. 2011;6(10):e26260. doi: 10.1371/journal.pone.0026260. Epub 2011 Oct 17. PLoS One. 2011. PMID: 22043312 Free PMC article. - High presence/absence gene variability in defense-related gene clusters of Cucumis melo.
González VM, Aventín N, Centeno E, Puigdomènech P. González VM, et al. BMC Genomics. 2013 Nov 12;14:782. doi: 10.1186/1471-2164-14-782. BMC Genomics. 2013. PMID: 24219589 Free PMC article. - Analysis of Extreme Phenotype Bulk Copy Number Variation (XP-CNV) Identified the Association of rp1 with Resistance to Goss's Wilt of Maize.
Hu Y, Ren J, Peng Z, Umana AA, Le H, Danilova T, Fu J, Wang H, Robertson A, Hulbert SH, White FF, Liu S. Hu Y, et al. Front Plant Sci. 2018 Feb 9;9:110. doi: 10.3389/fpls.2018.00110. eCollection 2018. Front Plant Sci. 2018. PMID: 29479358 Free PMC article. - Accurate prediction of quantitative traits with failed SNP calls in canola and maize.
Weber SE, Chawla HS, Ehrig L, Hickey LT, Frisch M, Snowdon RJ. Weber SE, et al. Front Plant Sci. 2023 Oct 23;14:1221750. doi: 10.3389/fpls.2023.1221750. eCollection 2023. Front Plant Sci. 2023. PMID: 37936929 Free PMC article. - Effector-Triggered Immune Response in Arabidopsis thaliana Is a Quantitative Trait.
Iakovidis M, Teixeira PJ, Exposito-Alonso M, Cowper MG, Law TF, Liu Q, Vu MC, Dang TM, Corwin JA, Weigel D, Dangl JL, Grant SR. Iakovidis M, et al. Genetics. 2016 Sep;204(1):337-53. doi: 10.1534/genetics.116.190678. Epub 2016 Jul 13. Genetics. 2016. PMID: 27412712 Free PMC article.
References
- Allen, R. L., P. Bittner-Eddy, L. Grenville-Briggs, J. Meitz, A. P. Rehmany et al., 2004. Host-parasite coevolutionary conflict between Arabidopsis and Downy Mildew. Science 306: 1957–1960. - PubMed
- Belkhadir, Y., R. Subramaiam and J. L. Dangl, 2004. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7: 391–399. - PubMed
- Bergelson, J., M. Kreitman, E. Stahl and D. Tian, 2001. Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. - PubMed
- Bittner-Eddy, P. D., L. R. Crute, E. B. Holub and J. L. Beynon, 2000. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21: 177–188. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous