Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer - PubMed (original) (raw)
Lisa M Hines, Nicole L Collins, Yanhui Hu, Ruwanthi N Gunawardane, Donna Moreira, Jacob Raphael, Daniel Jepson, Malvika Koundinya, Andreas Rolfs, Barbara Taron, Steven J Isakoff, Joan S Brugge, Joshua LaBaer
Affiliations
- PMID: 16512675
- PMCID: PMC2522320
- DOI: 10.1021/pr050395r
Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer
Abigail E Witt et al. J Proteome Res. 2006 Mar.
Abstract
Functional proteomics approaches that comprehensively evaluate the biological activities of human cDNAs may provide novel insights into disease pathogenesis. To systematically investigate the functional activity of cDNAs that have been implicated in breast carcinogenesis, we generated a collection of cDNAs relevant to breast cancer, the Breast Cancer 1000 (BC1000), and conducted screens to identify proteins that induce phenotypic changes that resemble events which occur during tumor initiation and progression. Genes were selected for this set using bioinformatics and data mining tools that identify genes associated with breast cancer. Greater than 1000 cDNAs were assembled and sequence verified with high-throughput recombination-based cloning. To our knowledge, the BC1000 represents the first publicly available sequence-validated human disease gene collection. The functional activity of a subset of the BC1000 collection was evaluated in cell-based assays that monitor changes in cell proliferation, migration, and morphogenesis in MCF-10A mammary epithelial cells expressing a variant of ErbB2 that can be inducibly activated through dimerization. Using this approach, we identified many cDNAs, encoding diverse classes of cellular proteins, that displayed activity in one or more of the assays, thus providing insights into a large set of cellular proteins capable of inducing functional alterations associated with breast cancer development.
Figures
Figure 1
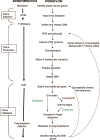
Schematic representation of workflow utilized to create the BC1000 collection.
Figure 2
Representative 3D structures of cells expressing cDNAs that scored as hits in the morphogenesis assays. MCF-10A cells expressing the pBABE control vector (pBABE) or EDG2, ARAF1, GRB2, JUNB, or BMP5 were cultured in morphogenesis assays as described in Materials and Methods for at least 16 days. The variability of morphogenic phenotypes among structures within the same experimental well is demonstrated by the inset in the BMP5 panel.
Figure 3
Representative 3D structures +/− ErbB2 dimerization, of 10A.B2 cells expressing cDNAs that scored in the sensitized migration assays. 10A.B2 cells were cultured with EGF in upper panels and with AP1510 in lower panels as described in Materials and Methods for at least 16 days.
Figure 4
Motile network structure of p16 and p21. Images taken six (upper) or 24 (lower) hours post-seeding on Matrigel, (A,D) MCF-10As plus EGF expressing vector control, (B,E) MCF-10A cells plus EGF expressing p16, and (C,F) MCF-10A cells plus EGF expressing p21.
Figure 5
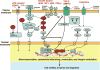
Genes identified in our screens in relation to various cellular pathways believed to be involved in motility, invasion and migration. Hits are in red.
References
- Nielsen KV, Niebuhr E, Ejlertsen B, Holstebroe S, Madsen MW, Briand P, Mouridsen HT, Bolund L. Molecular cytogenetic analysis of a nontumorigenic human breast epithelial cell line that eventually turns tumorigenic: validation of an analytical approach combining karyotyping, comparative genomic hybridization, chromosome painting, and single-locus fluorescence in situ hybridization. Genes Chromosomes Cancer. 1997;20:30–37. - PubMed
- Nishizaki T, DeVries S, Chew K, Goodson WH, 3rd, Ljung BM, Thor A, Waldman FM. Genetic alterations in primary breast cancers and their metastases: direct comparison using modified comparative genomic hybridization. Genes Chromosomes Cancer. 1997;19:267–272. - PubMed
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96:9212–9217. - PMC - PubMed
- Porter DA, Krop IE, Nasser S, Sgroi D, Kaelin CM, Marks JR, Riggins G, Polyak K. A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res. 2001;61:5697–5702. - PubMed
- Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–5979. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA089393/CA/NCI NIH HHS/United States
- P01 CA080111/CA/NCI NIH HHS/United States
- CA099191/CA/NCI NIH HHS/United States
- U54 GM64346-04/GM/NIGMS NIH HHS/United States
- CA080111/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous