Regulation of insulin receptor substrate 1 pleckstrin homology domain by protein kinase C: role of serine 24 phosphorylation - PubMed (original) (raw)
Regulation of insulin receptor substrate 1 pleckstrin homology domain by protein kinase C: role of serine 24 phosphorylation
Ranmali Nawaratne et al. Mol Endocrinol. 2006 Aug.
Abstract
Phosphorylation of insulin receptor substrate (IRS) proteins on serine residues is an important posttranslational modification that is linked to insulin resistance. Several phosphoserine sites on IRS1 have been identified; the majority are located proximal to the phosphotryosine-binding domain or near key receptor tyrosine kinase substrate- and/or Src-homology 2 domain-binding sites. Here we report on the characterization of a serine phosphorylation site in the N-terminal pleckstrin homology (PH) domain of IRS1. Bioinformatic tools identify serine 24 (Ser24) as a putative substrate site for the protein kinase C (PKC) family of serine kinases. We demonstrate that this site is indeed a bona fide substrate for conventional PKC. In vivo, IRS-1 is also phosphorylated on Ser24 after phorbol 12-myristate 13-acetate treatment of cells, and isoform-selective inhibitor studies suggest the involvement of PKCalpha. By comparing the pharmacological characteristics of phorbol 12-myristate 13-acetate-stimulated Ser24 phosphorylation with phosphorylation at two other sites previously linked to PKC activity (Ser307 and Ser612), we show that PKCalpha is likely to be directly involved in Ser24 phosphorylation, but indirectly involved in Ser307 and Ser612 phosphorylation. Using Ser24Asp IRS-1 mutants to mimic the phosphorylated residue, we demonstrate that the phosphorylation status of Ser24 does play an important role in regulating phosphoinositide binding to, and the intracellular localization of, the IRS1-PH domain, which can ultimately impinge on insulin-stimulated glucose uptake. Hence we provide evidence that IRS1-PH domain function is important for normal insulin signaling and is regulated by serine phosphorylation in a manner that could contribute to insulin resistance.
Figures
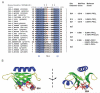
Fig. 1
Phosphorylation Potential, Conservation, and Structural Location of IRS-1 Serine 24 A, Sequence and structural alignment of N-terminal sequences of PH domains from all known IRS proteins and related phosphopotential scores. Sequence alignments were generated by ClustalW and compared with the known structure of hIRS1 (PDB:1qqga) using Fugue. Structural information for 1qqga is represented as follows: _β_-strand (blue), solvent accessible (lowercase), solvent inaccessible (uppercase), hydrogen bond to main-chain amide (bold), hydrogen bond to main-chain carbonyl (underline), and positive _ϕ_-torsion angle (italic). All numbering (except xIRS1) is according to Swissprot/Trembl. Sequence numbering for the xIRS1 sequence is taken from Ref. . Partial sequence availability is indicated by *. Arrows indicate key phospholipid-binding residues (26). Phosphopotential of each sequence was determined, using NetPhos and MotifScan (high stringency). Only one residue in the PH domain scored highly by both platforms (uppercase, bold, and red). This was Ser24 in rmhIRS1 (analogous to Ser14 in sIRS1, Ser27 in xIRS1, Ser44 in rIRS3, and Thr20 in dIRS1). Scores corresponding to each putative phospho residue are presented on the right. B, Ribbon diagrams of hIRS1 PH domain. _β_-Sheets are in _green, α_-helices in blue, and intervening coils or loops in yellow. The location of basic residues implicated in lipid binding (i.e. Lys 21, Lys 23, His 26, Arg 28, Lys 61, and Arg 62) are represented by red sticks and Ser24 by spheres. nd, Not detected.
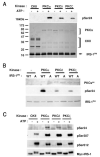
Fig. 2
In Vitro Phosphorylation of IRS1 PH Domain on Ser24 A and B, wt (WT) or S24A (A) recombinant His6-IRS-PH domains were incubated with recombinant PKC isoforms or casein kinase II (CKII) as described in Materials and Methods. The kinase reactions were terminated and proteins separated by SDS PAGE followed by Western blot analysis with an antibody against pSer24. Parallel gels were also run and stained with either Coomassie (lower panels in A and B) or silver stain (SS in panel B) to confirm equal loading of kinase and recombinant IRS-PH domains. PKC-mediated anti-pSer24 immunoreactivity was ATP dependent (panel A) and Ser24 specific (panel B). C, In vitro kinase reactions were performed as described for panel A except that substrate used here was full-length rIRS1 (Myc-tagged) immunoprecipitated from serum-starved NIH/IR-rIRS1wt-MycHis6 cell lysates. Proteins were separated by SDS-PAGE followed by sequential Western blot analysis with antibodies against pSer24, pSer307, pSer612, or myc. The membranes were stripped clean of primary antibodies between each Western blot. Data are representative of at least three independent experiments. CKII, Casein kinase II.
Fig. 3
In Vivo Phosphorylation of Full-Length rIRS1 on Serines 24, 307, and 612 NIH/hIR cells ectopically expressing either rIRS1wt or rIRS1 S24A were serum starved for 4 h and stimulated with either PMA (1 μ
m
), C2 ceramide (100 μ
m
), insulin (1 μ
m
) or dimethylsulfoxide (vehicle control) for 1.5 h. Monolayers were washed with ice-cold PBS, and total protein was extracted and quantified. IRS1 immunoprecipitates were collected from 2.5 mg protein lysates and separated by SDS-PAGE. Proteins were transferred to polyvinylidine difluoride membranes and sequentially immunoblotted using antibodies against pSer24, pSer307, pSer612, or myc. The membranes were stripped clean of primary antibodies between each Western blot. Cntrl, Control; Ins, insulin; C2, C2 ceramide.
Fig. 4
Effect of Selective PKC Inhibitors on PMA-Stimulated IRS1 Phosphorylation on Ser24, Ser307, and Ser612 A, Serum-starved NIH/hIR cells ectopically expressing rIRS1-wt were pretreated with indicated inhibitors for 30 min (or 24 h for chronic PMA) before stimulation with or without PMA (1 μ
m
). IRS1 immunoprecipitates were collected and analyzed as described for Fig. 3. Selectivities of inhibitors are as follows: Gö6976 (10 μ
m
) inhibits PKC α and β but not δ, ε, or ζ; Rottlerin (10 μ
m
) inhibits nPKCs δ and θ to a significantly greater extent than cPKCs α, β, and γ and has the least effect on PKCs ∊, η, and ζ; BAPTA-AM (10 μ
m
) inhibits all calcium-dependent cPKCs; calphostin (100 n
m
) inhibits all DAG-dependent PKCs (cPKC and nPKC); and Go6983 (100 n
m
) inhibits cPKC α and β but not PKC μ, δ, ∊, or ζ. Chronic PMA pretreatment down-regulated DAG-dependent PKCs α and δ but not μ and ζ. PKC β and γ were not detected (data not shown). Equal loading was confirmed by blotting against the p85 subunit of PI3K. Data are representative of at least three independent experiments. C, Rottlerin treatment reduced PMA-stimulated PKC_δ_ activation. Whole-cell lysates from experiments described in panel A were probed for active PKC_δ_ using anti-pSer643-PKC_δ_ (upper panel) or PKC_δ_ protein (lower panel).
Fig. 5
Effect of Serine Kinase Inhibitors on PMA-Stimulated IRS1 Phosphorylation on Ser24, Ser307, and Ser612 A, Serum-starved NIH/hIR cells ectopically expressing rIRS1-wt were pretreated with indicated inhibitors for 30 min and then stimulated with or without PMA (1 μ
m
). IRS1 immunoprecipitates were collected and analyzed as described for Fig. 3. The inhibitors used selectively target MEK1 (PD098059, 50 μ
m
), JNK (SP600125, 20 μ
m
), p38 (SB203580, 10 μ
m
), IKK_β_ (salicylic Acid, 5 m
m
), mTOR (rapamycin, 200 nm), GSK3 (LiCl2, 25 m
m
), and P13K (LY294002, 20 μ
m
). B, Schematic diagram illustrating PMA-stimulated serine kinases involved in site-specific serine phosphorylation of IRS1. Data are representative of at least three independent experiments.
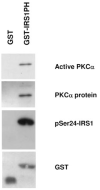
Fig. 6
Coprecipitation of IRS1 PH Domain with PKC_α_ Recombinant PKC_α_ and glutathione beads were incubated with either GST peptide alone or GST-IRS1-PH domain. In vitro kinase assays were performed on GST precipitates followed by SDS-PAGE and immunoblotting against pser24, active PKC_α_, GST, and PKC_α_.
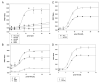
Fig. 7
Role of Serine 24 in Phosphoinositide Binding to IRS1 PH Domain A, Lipid binding to recombinant wt GST-IRS1-PH domain was determined as described in Materials and Methods using increasing concentrations of the indicated biotinylated phosphoinositide and determining binding by generation of a FRET signal. B, PtdIns(4,5)P2 binding to wt or mutant GST-IRS1-PH domains was determined as in panel A. FRET signals were normalized for absolute peptide concentration. C, Recombinant wt GST-IRS1-PH domain was incubated with or without recombinant PKC_ζ_ (minus cofactors) for 2 h at 30 C before determination of PtdIns(4,5)P2 binding. D, Recombinant wt GST-IRS1-PH domain was incubated with recombinant PKC_ζ_ (minus cofactors) in the presence or absence of ATP for 2 h at 30C before determination of PtdIns(4,5)P2 binding. Recombinant PKC_ζ_ was itself not tagged with GST. PI(3,4,5)P3, Phosphatidylinositol 3,4,5-triphosphate; PI(3,4)P2, phosphatidylinositol 3,4-bisphosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PI(3)P, phosphatidylinositol 3-monophosphate; PI(4)P, phosphatidylinositol 4-monophosphate.
Fig. 8
Intracellular Localization of GFP-Tagged PH Domain Mutants HEK293 cells were grown on poly-
l
-lysine-coated cover-slips and transiently transfected with either pNEGFP, pNEGFP-IRS1-PH wt, pNEGFP-IRS1-PH S24A, or pNEGFP-IRS1-PH S24D. Monolayers were serum starved (overnight) 48 h post transfection, washed with PBS, fixed in 2% formalin, and mounted in aqueous mount. Confocal fluorescent images were captured using a Zeiss imaging microscope set to capture GFP and DAPI blue fluorescence from a slice thickness of 0.8 nm using a ×60 oil emersion objective. Representative pictures are shown from at least three independent experiments performed in duplicate.
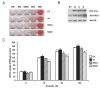
Fig. 9
3T3-L1 Adipocytes Expressing rIRS-1 Ser24 Mutants Show Similar Adipogenic Profiles and Levels of Mutant Expression but Respond Differently to Insulin-Stimulated Glucose Uptake A, 3T3-L1 preadipocytes were induced to differentiate with range of adipogenic treatments; adipocyte media only (AM), AM supplemented with insulin (INS), AM supplemented with isobutylmethylxanthine only (IBMX), AM supplemented with dexamethasone only (Dex), and AM supplemented with full induction cocktail of isobutylmethylxanthine, dexamthasone, and insulin (MDI). On d 8, these were fixed and accumulated lipids stained with oil red O. B, Equal amounts of total protein extract from mature adipocytes were separated on SDS-PAGE and immunoblotted for myc-tagged mutants, total IRS-1 protein, and IR_β_ as a marker of differentiation. C, Mature adipocytes expressing IRS-1 Ser24 mutants were serum starved (24 h) and 2-deoxy-
d
-[2,6 3H]-glucose uptake was determined in triplicate. Data represent mean ±
sd
and is representative of at least four independent experiments. Asterisk indicates statistical significance (P < 0.01) in comparison with wt cells with the same treatment. EV, Empty vector; DPM, disintegrations per min.
Similar articles
- Fatty acid-induced insulin resistance: role of insulin receptor substrate 1 serine phosphorylation in the retroregulation of insulin signalling.
Le Marchand-Brustel Y, Gual P, Grémeaux T, Gonzalez T, Barrès R, Tanti JF. Le Marchand-Brustel Y, et al. Biochem Soc Trans. 2003 Dec;31(Pt 6):1152-6. doi: 10.1042/bst0311152. Biochem Soc Trans. 2003. PMID: 14641015 Review. - Positive and negative regulation of insulin signaling through IRS-1 phosphorylation.
Gual P, Le Marchand-Brustel Y, Tanti JF. Gual P, et al. Biochimie. 2005 Jan;87(1):99-109. doi: 10.1016/j.biochi.2004.10.019. Biochimie. 2005. PMID: 15733744 Review.
Cited by
- Insulin and metabolic stress stimulate multisite serine/threonine phosphorylation of insulin receptor substrate 1 and inhibit tyrosine phosphorylation.
Hançer NJ, Qiu W, Cherella C, Li Y, Copps KD, White MF. Hançer NJ, et al. J Biol Chem. 2014 May 2;289(18):12467-84. doi: 10.1074/jbc.M114.554162. Epub 2014 Mar 20. J Biol Chem. 2014. PMID: 24652289 Free PMC article. - TNF-alpha and adipocyte biology.
Cawthorn WP, Sethi JK. Cawthorn WP, et al. FEBS Lett. 2008 Jan 9;582(1):117-31. doi: 10.1016/j.febslet.2007.11.051. Epub 2007 Nov 26. FEBS Lett. 2008. PMID: 18037376 Free PMC article. Review. - The Randle cycle revisited: a new head for an old hat.
Hue L, Taegtmeyer H. Hue L, et al. Am J Physiol Endocrinol Metab. 2009 Sep;297(3):E578-91. doi: 10.1152/ajpendo.00093.2009. Epub 2009 Jun 16. Am J Physiol Endocrinol Metab. 2009. PMID: 19531645 Free PMC article. Review. - 11beta-hydroxysteroid dehydrogenase type 1 regulates glucocorticoid-induced insulin resistance in skeletal muscle.
Morgan SA, Sherlock M, Gathercole LL, Lavery GG, Lenaghan C, Bujalska IJ, Laber D, Yu A, Convey G, Mayers R, Hegyi K, Sethi JK, Stewart PM, Smith DM, Tomlinson JW. Morgan SA, et al. Diabetes. 2009 Nov;58(11):2506-15. doi: 10.2337/db09-0525. Epub 2009 Aug 12. Diabetes. 2009. PMID: 19675138 Free PMC article. - Metabolic Messengers: tumour necrosis factor.
Sethi JK, Hotamisligil GS. Sethi JK, et al. Nat Metab. 2021 Oct;3(10):1302-1312. doi: 10.1038/s42255-021-00470-z. Epub 2021 Oct 14. Nat Metab. 2021. PMID: 34650277 Review.
References
- Zick Y. Uncoupling insulin signalling by serine/threonine phosphorylation: a molecular basis for insulin resistance. Biochem Soc Trans. 2004;32:812–816. - PubMed
- Lee YH, White MF. Insulin receptor substrate proteins and diabetes. Arch Pharm Res. 2004;27:361–370. - PubMed
- Voliovitch H, Schindler DG, Hadari YR, Taylor SI, Accili D, Zick Y. Tyrosine phosphorylation of insulin receptor substrate-1 in vivo depends upon the presence of its pleckstrin homology region. J Biol Chem. 1995;270:18083–18087. - PubMed
- Myers MG, Jr, Grammer TC, Brooks J, Glasheen EM, Wang LM, Sun XJ, Blenis J, Pierce JH, White MF. The pleckstrin homology domain in insulin receptor substrate-1 sensitizes insulin signaling. J Biol Chem. 1995;270:11715–11718. - PubMed
- White MF. IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab. 2002;283:E413–E422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous