Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression - PubMed (original) (raw)
Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression
Sandrine Tyteca et al. EMBO J. 2006.
Abstract
The histone acetyl transferase Tip60 (HTATIP) belongs to a multimolecular complex involved in the cellular response to DNA damage. Tip60 participates in cell cycle arrest following DNA damage by allowing p53 to activate p21CIP (p21) expression. We show here that Tip60 and the E1A-associated p400 protein (EP400), which belongs to the Tip60 complex, are also required for DNA damage-induced apoptosis. Tip60 favours the expression of some proapoptotic p53 target genes most likely through the stimulation of p53 DNA binding activity. In contrast, p400 represses p21 expression in unstressed cells, thereby allowing cell cycle progression and DNA damage-induced apoptosis. Tip60 and p400 have thus opposite effects on p21 expression in the absence of DNA damage. We further found that this antagonism relies on the inhibition of Tip60 function by p400, a property that is abolished following DNA damage. Therefore, taken together, our results indicate that Tip60 and p400 play distinct roles in DNA damage-induced apoptosis and underline the importance of the Tip60 complex and its regulation in the proper control of cell fate.
Figures
Figure 1
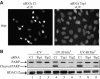
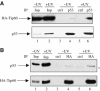
Tip60 is required for p53-dependent apoptosis. (A) U2OS cells were transfected by the indicated siRNAs using oligofectamine. After 48 h, they were UV-irradiated (80 J/m2), fixed with paraformaldehyde after 6 h and stained with dapi. (B) U2OS cells were transfected by the indicated siRNAs with oligofectamine. After 48 h, these cells were UV-irradiated (20 or 80 J/m2) or not, as indicated. After 4 h, Laemmli sample buffer was added to the transfected cells to ensure extraction of total proteins. Total cell extracts were then analysed for the presence of PARP (full-length or cleaved) and HDAC1/2 (as a loading control) by Western blot.
Figure 2
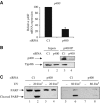
p400 and Tip49b are also required for UV-induced apoptosis. (A) U2OS cells were transfected with the indicated siRNAs using oligofectamine. After 48 h, they were harvested by scraping. Total RNAs were extracted and reverse transcribed. The amount of p400 cDNAs was measured by real-time PCR, divided by the amount of P0 cDNA and calculated relative to 100% for cells transfected with the control siRNAs. (B) Same as in (A), except that whole-cell extracts were prepared, immunoprecipitated with the anti-p400 antibody and blotted using anti-p400 or anti-Tip49b antibody. Also shown is a Western blot monitoring Tip49b levels in input material (p400 Western blots did not work properly on whole cell extracts). (C) U2OS cells were transfected with the indicated siRNAs using oligofectamine. After 48 h, the transfected cells were UV-irradiated (20 or 80 J/m2) or not. After 4 h, Laemmli sample buffer was added to the transfected cells to ensure extraction of total proteins. Cell extracts were then analysed for the presence of PARP by Western blot.
Figure 3
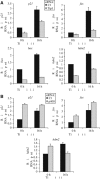
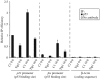
Tip60 and p400 have opposite effects on the expression of p53-dependent genes. (A) U2OS cells were transfected by the C1 siRNAs (black bars) or the Tip siRNAs (grey bars) by electroporation. After 48 h, they were UV-irradiated or not (10 J/m2), as indicated, and harvested 16 h later by scraping. Total RNAs were extracted and reverse transcribed. The amounts of p21, fas, bax and hdm2 cDNA were measured by Q-PCR, divided by the amount of ribosomal phosphoprotein P0 cDNA and calculated relative to 1 for cells transfected by the control siRNAs and not irradiated. A representative experiment is shown. (B) U2OS cells were transfected by the C1 siRNAs (black bars) or the p400 siRNAs (grey bars) by electroporation and treated as in (A). The amounts of p21, fas and hdm2 cDNA were measured by real time PCR and analysed as in (A). A representative experiment is shown.
Figure 4
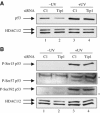
Tip60 knockdown does not affect p53 activation by UV-C. (A) U2OS cells were transfected by the indicated siRNAs and UV-irradiated (10 J/m2) 48 h later, where indicated. After 16 h, total cell extracts were prepared and analysed by Western blot for the presence of p53 (upper panel). After stripping, the membrane was blotted using an antibody recognising HDAC1/2 (lower panel). (B) Same as in (A), except that total cell extracts were analysed by Western blot for the presence of S15-, S37- and S392-phosphorylated p53. The stars indicate nonspecific bands.
Figure 5
Physical interaction between Tip60 and p53. (A) U2OS cells stably expressing HA-tagged Tip60 were UV-irradiated (10 J/m2) or not. After 16 h, total cell extracts were prepared and subjected to immunoprecipitation using an anti-p53 or an irrelevant antibody (ctrl) as indicated. Immunoprecipitated proteins were then analysed for the presence of p53 and Tip60-HA by Western blot. In total, 10% of inputs (Inp) were also loaded on the gel (lanes 1 and 2). (B) Same as in (A), except that cell extracts were immunoprecipitated by an anti-HA or a control antibody. The star indicates the migration of immunoglobulin heavy chains of the immunoprecipitating antibody, which is slightly detected by the secondary antibody (see e.g. in lane 5).
Figure 6
Tip60 is required for p53 binding on p53 target promoters. U2OS cells were transfected by the indicated siRNAs (C1 or Tip1) by electroporation and 48 h later, they were UV-irradiated or not (10 J/m2). After 16 h irradiation, ChIP was performed on these cells using an anti-p53 antibody (black bars) or no antibody as a control (grey bars). The presence of p21 promoter, fas promoter (containing the p53 response element) and β-actin coding sequence in the immunoprecipitates and in the inputs were analysed by real-time PCR. After dividing by the input DNA, the amount of immunoprecipitated sequence was calculated relative to 1 for the amount of p21 promoter immunoprecipitated by the anti-p53 antibody in cells transfected by the control siRNA and not irradiated. The mean and standard deviation from three entirely independent experiments are shown.
Figure 7
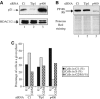
p400 knockdown induces a cell cycle arrest. (A) U2OS cells were transfected with the indicated siRNAs by electroporation. After 48 h, total cell extracts were prepared and analysed for the presence of p21 or HDAC (as a loading control) by Western blot. (B) Same as in (A), except that cell extracts were analysed for the presence of Rb by Western blot and for equal loading by ponceau red staining. (C) U2OS cells were transfected with the indicated siRNAs by electroporation. After 48 h later, the cell cycle distribution of transfected cells was analysed by flow cytometry.
Figure 8
p400 knockdown blocks apoptosis through p21 induction. (A, B) U2OS cells were transfected with the indicated siRNAs (10 μl of each siRNAs (100 μM)) by electroporation. The amount of siRNAs was kept constant using the control siRNA (C1). After 64 h, cells were harvested. Total RNAs were extracted and reverse transcribed. The amounts of p400, Tip60 (A) and p21 (B) cDNA were measured by real-time PCR and divided by the amount of P0 cDNA and standardised to 100% relative to the amount of cDNA in cells transfected by the C1 siRNA alone. A representative experiment is shown. (C) U2OS transfected as in (A) and (B) were harvested in Laemmli sample buffer and cell extracts were analysed for p21 protein expression by Western blot. (D) U2OS cells analysed in (A) and (B) were UV-irradiated or not (20 J/m2) and 4 h later, cell extracts were prepared and analysed for the presence of PARP by Western blot.
Figure 9
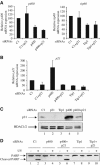
p400 functions as an inhibitor of Tip60 in unstressed cells. (A) U2OS cells were transfected with the indicated siRNAs (10 μl of each siRNAs (100 μM)) by electroporation. The amount of siRNAs was kept constant using the control siRNA (C1). After 64 h, cells were harvested. Total RNAs were extracted and reverse transcribed. The amount of p21 cDNA was measured by real-time PCR and divided by the amount of P0 cDNA and standardised to 100% relative to the amount of cDNA in cells transfected by the C1 siRNA alone. A representative experiment is shown. (B) U2OS cells transfected as in (A) were harvested in Laemmli sample buffer and cell extracts were analysed for Rb (top panel), p21 (middle panel) and HDAC (loading control, bottom panel) expression by Western blot. (C) U2OS cells were transfected with the indicated siRNAs as in (A). After 64 h, the cell cycle distribution of transfected cells was analysed by flow cytometry.
Similar articles
- The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways.
Mattera L, Escaffit F, Pillaire MJ, Selves J, Tyteca S, Hoffmann JS, Gourraud PA, Chevillard-Briet M, Cazaux C, Trouche D. Mattera L, et al. Oncogene. 2009 Mar 26;28(12):1506-17. doi: 10.1038/onc.2008.499. Epub 2009 Jan 26. Oncogene. 2009. PMID: 19169279 - Role of the histone acetyl transferase Tip60 in the p53 pathway.
Legube G, Linares LK, Tyteca S, Caron C, Scheffner M, Chevillard-Briet M, Trouche D. Legube G, et al. J Biol Chem. 2004 Oct 22;279(43):44825-33. doi: 10.1074/jbc.M407478200. Epub 2004 Aug 13. J Biol Chem. 2004. PMID: 15310756 - The SANT domain of p400 ATPase represses acetyltransferase activity and coactivator function of TIP60 in basal p21 gene expression.
Park JH, Sun XJ, Roeder RG. Park JH, et al. Mol Cell Biol. 2010 Jun;30(11):2750-61. doi: 10.1128/MCB.00804-09. Epub 2010 Mar 29. Mol Cell Biol. 2010. PMID: 20351180 Free PMC article. - Cellular functions of TIP60.
Sapountzi V, Logan IR, Robson CN. Sapountzi V, et al. Int J Biochem Cell Biol. 2006;38(9):1496-509. doi: 10.1016/j.biocel.2006.03.003. Epub 2006 Mar 28. Int J Biochem Cell Biol. 2006. PMID: 16698308 Review. - Tip60 in DNA damage response and growth control: many tricks in one HAT.
Squatrito M, Gorrini C, Amati B. Squatrito M, et al. Trends Cell Biol. 2006 Sep;16(9):433-42. doi: 10.1016/j.tcb.2006.07.007. Epub 2006 Aug 9. Trends Cell Biol. 2006. PMID: 16904321 Review.
Cited by
- RVBs are required for assembling a functional TIP60 complex.
Jha S, Gupta A, Dar A, Dutta A. Jha S, et al. Mol Cell Biol. 2013 Mar;33(6):1164-74. doi: 10.1128/MCB.01567-12. Epub 2013 Jan 7. Mol Cell Biol. 2013. PMID: 23297341 Free PMC article. - Evaluating the Cellular Roles of the Lysine Acetyltransferase Tip60 in Cancer: A Multi-Action Molecular Target for Precision Oncology.
Zohourian N, Coll E, Dever M, Sheahan A, Burns-Lane P, Brown JAL. Zohourian N, et al. Cancers (Basel). 2024 Jul 27;16(15):2677. doi: 10.3390/cancers16152677. Cancers (Basel). 2024. PMID: 39123405 Free PMC article. Review. - Activation of p21 by HDAC inhibitors requires acetylation of H2A.Z.
Bellucci L, Dalvai M, Kocanova S, Moutahir F, Bystricky K. Bellucci L, et al. PLoS One. 2013;8(1):e54102. doi: 10.1371/journal.pone.0054102. Epub 2013 Jan 18. PLoS One. 2013. PMID: 23349794 Free PMC article. - Experimental Insights into the Interplay between Histone Modifiers and p53 in Regulating Gene Expression.
Kim HM, Zheng X, Lee E. Kim HM, et al. Int J Mol Sci. 2023 Jul 3;24(13):11032. doi: 10.3390/ijms241311032. Int J Mol Sci. 2023. PMID: 37446210 Free PMC article. Review. - C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts.
Bortolin-Cavaillé ML, Dance M, Weber M, Cavaillé J. Bortolin-Cavaillé ML, et al. Nucleic Acids Res. 2009 Jun;37(10):3464-73. doi: 10.1093/nar/gkp205. Epub 2009 Apr 1. Nucleic Acids Res. 2009. PMID: 19339516 Free PMC article.
References
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG (2002) Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell 110: 55–67 - PubMed
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL (2001) Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 8: 1243–1254 - PubMed
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428: 431–437 - PubMed
- Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, Robson CN (1999) Tip60 is a nuclear hormone receptor coactivator. J Biol Chem 274: 17599–17604 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous