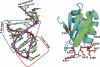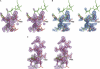The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold - PubMed (original) (raw)
The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold
Shuying Wang et al. RNA. 2006 Jun.
Abstract
The YxiN protein of Bacillus subtilis is a member of the DbpA subfamily of prokaryotic DEAD-box RNA helicases. Like DbpA, it binds with high affinity and specificity to segments of 23S ribosomal RNA as short as 32 nucleotides (nt) that include hairpin 92. Several experiments have shown that the 76-residue carboxy-terminal domain of YxiN is responsible for the high-affinity RNA binding. The domain has been crystallized and its structure has been solved to 1.7 Angstroms resolution. The structure reveals an RNA recognition motif (RRM) fold that is found in many eukaryotic RNA binding proteins; the RRM fold was not apparent from the amino acid sequence. The domain has two solvent exposed aromatic residues at sites that correspond to the aromatic residues of the ribonucleoprotein (RNP) motifs RNP1 and RNP2 that are essential for RNA binding in many RRMs. However, mutagenesis of these residues (Tyr404 and Tyr447) to alanine has little effect on RNA affinity, suggesting that the YxiN domain binds target RNAs in a manner that differs from the binding mode commonly found in many eukaryotic RRMs.
Figures
FIGURE 1.
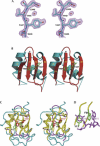
Structure of the YxiN RBD. (A) Experimental solvent-flipped electron density map contoured at 1.8σ. Molecular model shown for reference. (B) Ribbon drawing of the RBD structure. Dotted lines indicate segments of polypeptide that could not be traced. The side chains of Tyr407 and Tyr447 are also shown. (C) Superposition of the U1A RNA binding domain (PDB ID 1URN) and YxiN RBD, represented as a coil passing through Cα positions. Segments of polypeptide for which the distance between equivalent Cα’s of the two structures is <1.5 Å are shown in yellow; segments for which the distance is >1.5 Å, in magenta (YxiN) and cyan (U1A). Side chains of YxiN residues Tyr407 and Tyr447 are shown in red; side chains of the equivalent U1A residues Tyr13 and Phe56, in green. (D) Manner in which RNA nucleotide bases stack on aromatic side chains of U1A. RNA nucleotides C10 and A11, magenta; U1A Tyr13 and Phe56 side chains, green. A, C, D, and Figure 3 were made with Pymol (
http://pymol.sourceforge.net/index.php
); B was made with Molscript (Kraulis 1991) and rendered with Raster3D (Merritt and Bacon 1997).
FIGURE 2.
Binding of the YxiN RBD to RNA. (A) RNA 32-mer oligonucleotides used for binding measurements. RNA A is the wild-type sequence derived from 23S RNA, which includes hairpin H92. Mutations in oligonucleotides B–D are shown with large outline characters. (B) Representative gel shift assay data showing the binding of wild-type and Y447A mutant RBD proteins to RNA B. (C) Representative binding curves, plotting the binding of YxiN RBD wild-type (filled symbols) and Y447A mutant (open symbols) protein to RNA B (circles) and RNA D (triangles).
FIGURE 3.
YxiN RBD with conserved residues highlighted. (Right) Ribbon drawing on YxiN RBD, with residues shown that have conservative substitutions in 36 aligned sequences (Karginov et al. 2005). Conserved external residues, with side chains presented as stick models in red: K412, K413, K415, R417, D420, F/Y447, K468, K470. Internal residues having only conservative substitution (L/I/V), with side chains shown as green stick model surrounded by semitransparent surface: V422, I425, V431, I436, I439, I441, V448, I450, V467. Conserved glycines, shown as magenta spheres at Cα positions: G411, G423, G437, G469. Residues that are included in figure in an approximate conformation that follows continuous electron density but not in final model due to inability to precisely define their conformation: 415–418 and 468–470. Selected residues are labeled. (Left) For reference, an anticodon fragment of tRNA (taken from PDB 1EIY) having the same features of secondary structure as the tight-binding RNA target of the YxiN RBD: a hairpin with a 5-bp stem and a 5-nt loop, plus a single-strand extension.
FIGURE 4.
Electron density maps in region of helix α1, which displays conformational heterogeneity. Stick models for part of helix in unique conformation (residues 424–427, top of figures) colored green. Segment of helix built in two conformations (residues 418–423): alternate conformation 1 (AC1), green; AC2, red. The break point in the alternate conformations is residue Gly423. (A) Fo − Fc map computed with only AC2 included in calculation of model phases, contoured at 2.5σ. (B) Fo − Fc map computed with only AC1 included in calculation of model phases, contoured at 2.5σ. (C) 2_Fo_ − Fc simulated annealing omit map, contoured at 1.2σ. For clarity, orientation of the model in C differs slightly from that of A and B. Figure made with Pymol (
http://pymol.sourceforge.net/index.php
).
Similar articles
- The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA.
Kossen K, Karginov FV, Uhlenbeck OC. Kossen K, et al. J Mol Biol. 2002 Dec 6;324(4):625-36. doi: 10.1016/s0022-2836(02)01140-3. J Mol Biol. 2002. PMID: 12460566 - Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif.
Hardin JW, Hu YX, McKay DB. Hardin JW, et al. J Mol Biol. 2010 Sep 17;402(2):412-27. doi: 10.1016/j.jmb.2010.07.040. Epub 2010 Jul 29. J Mol Biol. 2010. PMID: 20673833 Free PMC article. - RNA-protein complexes.
Nagai K. Nagai K. Curr Opin Struct Biol. 1996 Feb;6(1):53-61. doi: 10.1016/s0959-440x(96)80095-9. Curr Opin Struct Biol. 1996. PMID: 8696973 Review. - RNA sculpting by the primordial Helix-clasp-Helix-Strand-Loop (HcH-SL) motif enforces chemical recognition enabling diverse KH domain functions.
Tainer JA, Tsutakawa SE. Tainer JA, et al. J Biol Chem. 2025 May;301(5):108474. doi: 10.1016/j.jbc.2025.108474. Epub 2025 Apr 2. J Biol Chem. 2025. PMID: 40185232 Free PMC article. Review.
Cited by
- The Thermus thermophilus DEAD box helicase Hera contains a modified RNA recognition motif domain loosely connected to the helicase core.
Rudolph MG, Klostermeier D. Rudolph MG, et al. RNA. 2009 Nov;15(11):1993-2001. doi: 10.1261/rna.1820009. Epub 2009 Aug 26. RNA. 2009. PMID: 19710183 Free PMC article. - A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit.
Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. Sharpe Elles LM, et al. Nucleic Acids Res. 2009 Oct;37(19):6503-14. doi: 10.1093/nar/gkp711. Epub 2009 Sep 4. Nucleic Acids Res. 2009. PMID: 19734347 Free PMC article. - Roles of DEAD-box proteins in RNA and RNP Folding.
Pan C, Russell R. Pan C, et al. RNA Biol. 2010 Nov-Dec;7(6):667-76. doi: 10.4161/rna.7.6.13571. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045543 Free PMC article. Review. - Contribution of teg49 small RNA in the 5' upstream transcriptional region of sarA to virulence in Staphylococcus aureus.
Kim S, Reyes D, Beaume M, Francois P, Cheung A. Kim S, et al. Infect Immun. 2014 Oct;82(10):4369-79. doi: 10.1128/IAI.02002-14. Epub 2014 Aug 4. Infect Immun. 2014. PMID: 25092913 Free PMC article. - Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA.
Grohman JK, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. Grohman JK, et al. Biochemistry. 2007 Mar 20;46(11):3013-22. doi: 10.1021/bi0619472. Epub 2007 Feb 21. Biochemistry. 2007. PMID: 17311413 Free PMC article.
References
- Allain F.H., Gilbert D.E., Bouvet P., Feigon J. Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J. Mol. Biol. 2000b;303:227–241. - PubMed
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. - PubMed
- Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous