Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells - PubMed (original) (raw)
Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells
Ranajoy Chattopadhyay et al. Nucleic Acids Res. 2006.
Abstract
Abasic (AP)-endonuclease (APE) is responsible for repair of AP sites, and single-strand DNA breaks with 3' blocking groups that are generated either spontaneously or during repair of damaged or abnormal bases via the DNA base excision repair (BER) pathway in both nucleus and mitochondria. Mammalian cells express only one nuclear APE, 36 kDa APE1, which is essential for survival. Mammalian mitochondrial (mt) BER enzymes other than mtAPE have been characterized. In order to identify and characterize mtAPE, we purified the APE activity from beef liver mitochondria to near homogeneity, and showed that the mtAPE which has 3-fold higher specific activity relative to APE1 is derived from the latter with deletion of 33 N-terminal residues which contain the nuclear localization signal. The mtAPE-sized product could be generated by incubating 35S-labeled APE1 with crude mitochondrial extract, but not with cytosolic or nuclear extract, suggesting that cleavage of APE1 by a specific mitochondria-associated N-terminal peptidase is a prerequisite for mitochondrial import. The low abundance of mtAPE, particularly in cultured cells might be the reason for its earlier lack of detection by western analysis.
Figures
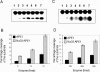
Figure 1
APE activity of Heparin–Sepharose fractions. (A) APE activity of fractions was measured at 37°C for 10 min with 1:200 diluted fractions and 500 nM substrate. (B) Quantitative representation of APE activity in active fractions (10–16) after 1:500 dilution.
Figure 2
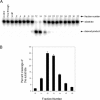
Identification of purified bovine mtAPE. (A) Coomassie staining of fraction 12 (25 µl; Figure 1). Lane 1, Recombinant NΔ33 hAPE1. Lane 2, fraction 12; M, molecular weight markers. (B) Western blot of fraction 12 with APE1 antibody. Lane 1, 20 ng recombinant hAPE1; Lane 2, fraction 12 (2 µl); Lane 3, recombinant NΔ20 APE1 (25 ng).
Figure 3
Confirmation of mtAPE as NΔ33 APE1. (A) MS analysis of trypsin-digested mtAPE band (fraction 12). (B) N-terminal sequence of mtAPE of EKEAV (shown in the box).
Figure 4
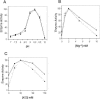
Kinetic parameters for recombinant hAPE1 (closed square) and bovine mtAPE (closed triangle). Dependence on (A) pH, (B) Mg2+ and (C) KCl.
Figure 5
Comparative endonuclease activity of recombinant hAPE1 and bovine mtAPE (fraction 12). (A) Quantification of hAPE1 and bovine mtAPE from western analysis. Lanes 1–3, 4, 8 and 16 ng of APE1, respectively; lanes 4–6, 0.5, 1 and 2 µl of fraction 12, respectively. (B) THF-oligo substrate was incubated with hAPE1 or fraction 12 as described in Materials and Methods. Lane 1, no enzyme; lane 2, 1 fmol hAPE1; lane 3, 0.5 fmol hAPE1; lanes 4–5, fraction 12, at 1000- and 2500-fold dilution, respectively.
Figure 6
Relative activity of NΔ33 and full-length hAPE1. (A) Endonuclease activity, lane 1, control; lanes 2–4, 0.1, 0.2 and 0.5 fmol full-length APE1; lanes 5–7, 0.1, 0.2 and 0.5 fmol NΔ33 APE1. (C) 3′ Phosphodiesterase activity, lane 1, control; lanes 2, 4 and 6; 100, 200 and 300 fmol APE1; lanes 3, 5and 7; 100, 200 and 300 fmol NΔ33 APE1. (B and D) Graphical representation of the results in (A) and (C), respectively.
Figure 7
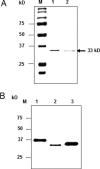
Presence of NΔ33 APE1 in beef liver mitochondria. (A) Lane 1, full-length and NΔ33 APE1 markers; lane 2, mitochondrial extract (50 µg); lane 3, extract (50 µg) of sucrose density gradient-purified mitochondria; lane 4, 25 µg extract after trypsin treatment for 20 min. (B) Lane 1, APE1 and NΔ33 APE1 markers; lanes 2–4, extract (25 µg) of nucleus, cytoplasm and mitochondria, respectively.
Figure 8
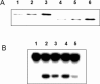
Presence of NΔ33 APE1 in NIH3T3 cells. (A) Western analysis. Lane 1, recombinant full length hAPE1; lane 2, 20 µg nuclear extract; lane 3, 20 µg cytoplasmic fraction; lane 4, 50 µg mitochondrial extract; lane 5, 50 µg mitochondrial extract after trypsin treatment for 20 min; lane 6, recombinant NΔ33 APE1. (B) Specific cleavage of full-length 35S-labeled hAPE1 by mitochondrial extract. Lane 1, APE1 control; lanes 2–4, treatment with mitochondrial, nuclear and cytoplasmic extracts, respectively as described in Materials and Methods. Full-length and cleaved products in duplicates (a and b) are indicated by arrows.
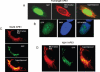
Figure 9
Intracellular localization of full-length and truncated hAPE1 with C-terminal EGFP tag. (A) Nuclear as well as cytoplasmic and mitochondrial localization of full-length APE1-EGFP in live cells. (B) Nuclear localization of full-length APE1 in fixed cells with nuclear DAPI staining. (C and D) Colocalization of NΔ20 APE1-EGFP and NΔ41 APE1-EGFP with MitoTracker Red showing their presence in the mitochondria.
Similar articles
- Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein.
Mitra S, Izumi T, Boldogh I, Bhakat KK, Chattopadhyay R, Szczesny B. Mitra S, et al. DNA Repair (Amst). 2007 Apr 1;6(4):461-9. doi: 10.1016/j.dnarep.2006.10.010. Epub 2006 Dec 12. DNA Repair (Amst). 2007. PMID: 17166779 - Long patch base excision repair in mammalian mitochondrial genomes.
Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Szczesny B, et al. J Biol Chem. 2008 Sep 26;283(39):26349-56. doi: 10.1074/jbc.M803491200. Epub 2008 Jul 17. J Biol Chem. 2008. PMID: 18635552 Free PMC article. - Modulation of the Apurinic/Apyrimidinic Endonuclease Activity of Human APE1 and of Its Natural Polymorphic Variants by Base Excision Repair Proteins.
Kladova OA, Alekseeva IV, Saparbaev M, Fedorova OS, Kuznetsov NA. Kladova OA, et al. Int J Mol Sci. 2020 Sep 28;21(19):7147. doi: 10.3390/ijms21197147. Int J Mol Sci. 2020. PMID: 32998246 Free PMC article. - Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.
McNeill DR, Whitaker AM, Stark WJ, Illuzzi JL, McKinnon PJ, Freudenthal BD, Wilson DM 3rd. McNeill DR, et al. Mutagenesis. 2020 Feb 13;35(1):27-38. doi: 10.1093/mutage/gez046. Mutagenesis. 2020. PMID: 31816044 Free PMC article. Review. - Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means.
Mol CD, Hosfield DJ, Tainer JA. Mol CD, et al. Mutat Res. 2000 Aug 30;460(3-4):211-29. doi: 10.1016/s0921-8777(00)00028-8. Mutat Res. 2000. PMID: 10946230 Review.
Cited by
- Human apurinic/apyrimidinic endonuclease 1.
Li M, Wilson DM 3rd. Li M, et al. Antioxid Redox Signal. 2014 Feb 1;20(4):678-707. doi: 10.1089/ars.2013.5492. Epub 2013 Aug 20. Antioxid Redox Signal. 2014. PMID: 23834463 Free PMC article. Review. - Mitochondrial DNA degradation: A quality control measure for mitochondrial genome maintenance and stress response.
Zhao L. Zhao L. Enzymes. 2019;45:311-341. doi: 10.1016/bs.enz.2019.08.004. Enzymes. 2019. PMID: 31627882 Free PMC article. Review. - NEIL1 and NEIL2 DNA glycosylases protect neural crest development against mitochondrial oxidative stress.
Han D, Schomacher L, Schüle KM, Mallick M, Musheev MU, Karaulanov E, Krebs L, von Seggern A, Niehrs C. Han D, et al. Elife. 2019 Sep 30;8:e49044. doi: 10.7554/eLife.49044. Elife. 2019. PMID: 31566562 Free PMC article. - Mechanism of hypoxia-inducible factor 1 alpha-mediated Mcl1 regulation in Helicobacter pylori-infected human gastric epithelium.
Bhattacharyya A, Chattopadhyay R, Hall EH, Mebrahtu ST, Ernst PB, Crowe SE. Bhattacharyya A, et al. Am J Physiol Gastrointest Liver Physiol. 2010 Nov;299(5):G1177-86. doi: 10.1152/ajpgi.00372.2010. Epub 2010 Sep 9. Am J Physiol Gastrointest Liver Physiol. 2010. PMID: 20829524 Free PMC article. - The Good, the Bad, and the Ugly of ROS: New Insights on Aging and Aging-Related Diseases from Eukaryotic and Prokaryotic Model Organisms.
Santos AL, Sinha S, Lindner AB. Santos AL, et al. Oxid Med Cell Longev. 2018 Mar 18;2018:1941285. doi: 10.1155/2018/1941285. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 29743972 Free PMC article. Review.
References
- Dodson M.L., Michaels M.L., Lloyd R.S. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 1994;269:32709–32712. - PubMed
- Giloni L., Takeshita M., Johnson F., Iden C., Grollman A.P. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J. Biol. Chem. 1981;256:8608–8615. - PubMed
- Henner W.D., Grunberg S.M., Haseltine W.A. Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J. Biol. Chem. 1983;258:15198–15205. - PubMed
- Demple B., Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG021830/AG/NIA NIH HHS/United States
- R01 CA098664/CA/NCI NIH HHS/United States
- ES08457/ES/NIEHS NIH HHS/United States
- R01 ES008457/ES/NIEHS NIH HHS/United States
- R01 CA053791/CA/NCI NIH HHS/United States
- CA98664/CA/NCI NIH HHS/United States
- P30 ES006676/ES/NIEHS NIH HHS/United States
- ES06676/ES/NIEHS NIH HHS/United States
- R56 CA098664/CA/NCI NIH HHS/United States
- R01 CA53791/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous