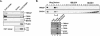GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation - PubMed (original) (raw)
GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation
Jeong Hyeon Park et al. Mol Cell Biol. 2006 Jun.
Abstract
GAS41 is a common subunit of the TIP60 and SRCAP complexes and is essential for cell growth and viability. Here, we report that GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Either GAS41 small interfering RNA-mediated knockdown of GAS41 expression or specific interruptions of the carboxy-terminal coiled-coil motif of the GAS41 protein activate the p53 tumor suppressor pathway, as evidenced by p53 up-regulation, p53 serine-15 phosphorylation, and p21 transcriptional activation. Activation of the p53 pathway does not result from changes in TIP60 complex assembly or TIP60 coactivator functions for p53, since a TIP60 complex containing a coiled-coil mutant of GAS41 retains the same composition and histone acetyltransferase activity as its wild-type counterpart and since mutant GAS41 does not compromise ectopic p53-dependent transcriptional activation in a reporter gene assay. Finally, we demonstrate that GAS41 is prebound to the promoters of two p53 tumor suppressor pathway genes (p21 and p14ARF) in normal unstressed cells but is dissociated from both promoters in response to stress signals that activate p53. Our data suggest that GAS41 plays a role in repressing the p53 tumor suppressor pathway during the normal cell cycle by a TIP60-independent mechanism.
Figures
FIG. 1.
GAS41 is a core subunit of the human TIP60 and SRCAP complexes. (A) Coimmunoprecipitation of GAS41 with core subunits of the TIP60 complex. 293T cells were transfected with empty vector or vector expressing FLAG-GAS41, and whole-cell lysates were immunoprecipitated with M2 agarose beads. After extensive washes, the captured FLAG-GAS41 was eluted with FLAG peptide under nondenaturing conditions. Eluates were analyzed by immunoblotting and HAT assays as indicated. Histone H2A was not separated from H3 in this minigel in which acetylated H2A partially overlapped with H3 (see Fig. 4B for better separation). (B) Sepharose CL6B gel filtration chromatography. Nuclear extracts were prepared from 293T cells stably expressing FLAG-GAS41 and directly applied to the Sepharose CL6B gel filtration column. Each fraction (0.1%) was analyzed by immunoblotting for TRRAP and GAS41. Fractions corresponding to the TRRAP peak (fractions 4 and 5) were combined and subjected to M2 agarose affinity purification. After being washed, captured FLAG-GAS41 was sequentially eluted with FLAG peptide and 0.1 M glycine (pH 2.5) and analyzed by immunoblotting for TRRAP, TIP60, p400, H2AZ, and GAS41.
FIG. 2.
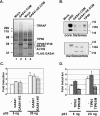
Effects of GAS41 siRNA on p53-dependent p21 gene induction and cell cycle progression. (A) GAS41 siRNA-mediated GAS41 knockdown in U2OS cells. Cells were transfected in duplicate with the indicated siRNAs, and whole-cell lysates were analyzed for relative levels of GAS41 and BAF53. (B) GAS41 siRNA-mediated induction of the p21 gene in U2OS cells. Cells were transfected with the indicated siRNAs and, after 48 h, treated with actinomycin D (10 ng/ml) for 6 h as indicated. Total RNA was analyzed by RT-PCR to estimate the relative expression levels of the p21 and GAPDH mRNAs. Relative levels of p21 gene expression (normalized to GAPDH expression) are shown in italics. (C) Effects of GAS41 siRNA on the levels of GAS41 and p53 in U2OS cells. Cells were transfected with the indicated siRNAs and, after 48 h, treated with actinomycin D (10 ng/ml) for 6 h as indicated. Total cell lysates were analyzed by immunoblotting for GAS41, p53, and actin. (D) Effects of GAS41 siRNA on cell cycle progression. U2OS cells were transfected in duplicate with the indicated siRNAs for 48 h and analyzed for DNA content by fluorescence-activated cell sorter analysis. Relative distributions of cell cycle were obtained from the analysis of FlowJo software. (E) Failure of GAS41 siRNA to induce p21 gene expression in p53-deficient H1299 cells. H1299 cells were transfected with control or GAS41 siRNA and, after 24 h, with empty vector (1 μg [−]) or vector expressing p53 (1 [+] or 2 [++] μg) as indicated. Total RNA was analyzed by RT-PCR for p21 and GAPDH mRNAs. (F) Effects of GAS41 siRNA on the levels of GAS41 and ectopically expressed p53 in H1299 cells. Cells were transfected with control or GAS41 siRNA and, after 24 h, with empty vector (1 μg [−]) or vector expressing p53 (1 [+] or 2 [++] μg). After 48 h, total cell lysates were analyzed by immunoblotting for GAS41, p53, and actin.
FIG. 3.
Structure-function analysis of the GAS41 coiled-coil motif. (A) Schematic diagram of four heptad repeats in the coiled-coil domain in the GAS41 and YAF9 C termini. The mutated amino acid residues are indicated by position numbers. (B) Disruption of the coiled-coil structure induces p21 gene activation. U2OS cells were transiently transfected with an empty vector (V) or a vector expressing wild-type (WT) or mutant GAS41. At 24 h posttransfection, total RNA and whole-cell extracts were analyzed by RT-PCR and immunoblotting, respectively. (C) Requirement of the GAS41 coiled-coil structure for interaction with the TIP60 complex. 293T cells were transfected with an empty vector or a vector expressing FLAG-GAS41, and whole-cell lysates were immunoprecipitated with M2 agarose beads. Eluates were analyzed by immunoblotting and HAT assays as indicated. (D) Requirement of the GAS41 coiled-coil structure for interaction with p400 and H2AZ. An experiment equivalent to that used for panel C was conducted to analyze interactions of GAS41 mutants with p400 and H2AZ by coimmunoprecipitation. (E) Activation of the p53 tumor suppressor pathway both by GAS41 mutants that are defective in TIP60 and SRCAP complex assembly and by GAS41 mutants that are not. U2OS cells were transiently transfected with the same set of GAS41 mutant vectors as those shown in panel C, and whole-cell lysates were analyzed by immunoblotting as indicated. Cells transfected with an empty vector followed by actinomycin D treatment (10 ng/ml) for 6 h were used as a positive control for activation of the p53 pathway. An immunoblot against BAF53 was used as a loading control.
FIG. 4.
Biochemical analysis of a GAS41 mutant-containing TIP60 complex. (A) Two-step purification of the GAS41-containing TIP60 complex. Nuclear extracts of 293T cells stably expressing FLAG-GAS41 or the FLAG-GAS41 (K212A/E214A) mutant were first fractionated by Sepharose CL6B gel filtration. Fractions corresponding to the TRRAP peak were combined and subjected to M2 agarose affinity purification. After being washed, captured FLAG-GAS41 was eluted with FLAG peptide and analyzed by silver staining. A mock purification was also conducted with a parental 293T cell extract using the same procedures. (B) HAT activities of the GAS41 mutant-containing TIP60 complex on free core histones and nucleosomes. Equivalent amounts of purified complexes containing GAS41 or the GAS41 (K212A/E214A) mutant proteins (A) were assayed for HAT activity using HeLa core histones or in vitro-assembled chromatin. Recombinant p300 (from baculovirus) and TIP60 (from Escherichia coli) were used as controls. (C) Effects of a GAS41 mutant on p53-dependent transcriptional activation. H1299 cells were transfected with pVP-p53 (5 ng or 20 ng), a p21 promoter-driven luciferase plasmid, and 200 ng of empty vector (−) or vector expressing wild-type GAS41 or the GAS41 (K212A/E214A) mutant. Inductions (_n_-fold) were calculated by dividing the normalized p53-dependent luciferase activity by the basal activity observed in the absence of p53 expression. The data presented are average values from triplicate samples. (D) Effects of a catalytically null TIP60 mutant on p53-dependent transcriptional activation. H1299 cells were transfected with pVP-p53 (5 ng or 20 ng), a p21 promoter-driven luciferase plasmid, and 200 ng of empty vector (−) or vector expressing wild-type TIP60 or the TIP60M (Q377E) mutant. Inductions (_n_-fold) were calculated by dividing the normalized p53-dependent luciferase activity by the basal activity observed in the absence of p53 expression. The data presented are average values from triplicate samples.
FIG. 5.
GAS41 binds to the promoters of repressed genes in the p53 tumor suppressor pathway. (A) Induction of p14 ARF gene expression by a coiled-coil mutant of GAS41. Retroviruses made from empty vector (LXSH) or from vectors expressing wild-type GAS41 or the GAS41 (K212A/E214A) mutant were used to infect U2OS and primary IMR-90 cells, and infected cells were selected for 7 days in hygromycin. Total RNA was purified and analyzed by RT-PCR for relative levels of p14 ARF and GAPDH mRNAs. Total cell lysates were analyzed by immunoblotting for relative levels of p53, FLAG-GAS41, and actin. (B) Binding of GAS41 to the p14 ARF gene promoter in unstressed IMR-90 cells. A ChIP assay was performed with anti-GAS41 antibody using proliferating IMR-90 cells. Final eluates were analyzed by PCR against the E2F binding region of the p14 ARF gene promoter. As a background control, the β-globin intron region was amplified in the same PCR. (C) Dissociation of prebound GAS41 from the p14 ARF promoter upon gene activation. IMR-90 cells were treated in the presence or absence of 20 μM roscovitine for 6 h, and ChIP assays were performed for the p14 ARF gene promoter and β_-_globin intron regions using antibodies against the H2AZ histone variant (as a control) and GAS41. (D) Differential binding of TIP60 and GAS41 to p21 and p14 ARF gene promoters. U2OS cells and H1299 cells were grown in the presence or absence of actinomycin D (10 ng/ml) for 6 h, and ChIP assays were performed for the p21 and p14 ARF gene promoter regions using antibodies as indicated. (E) Binding of the GAS41 (K212A/E214A) mutant to the p21 and p14 ARF gene promoters. Retroviruses expressing wild-type FLAG-GAS41 or the FLAG-GAS41 (K212A/E214A) mutant were used to infect primary IMR-90 cells. After 7 days of hygromycin selection, infected cells were used for the ChIP assay using anti-FLAG antibody.
Similar articles
- Regulation of Cell Proliferation and Migration by miR-203 via GAS41/miR-10b Axis in Human Glioblastoma Cells.
Pal D, Mukhopadhyay D, Ramaiah MJ, Sarma P, Bhadra U, Bhadra MP. Pal D, et al. PLoS One. 2016 Jul 28;11(7):e0159092. doi: 10.1371/journal.pone.0159092. eCollection 2016. PLoS One. 2016. PMID: 27467502 Free PMC article. - Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics.
Aslanian A, Iaquinta PJ, Verona R, Lees JA. Aslanian A, et al. Genes Dev. 2004 Jun 15;18(12):1413-22. doi: 10.1101/gad.1196704. Epub 2004 Jun 2. Genes Dev. 2004. PMID: 15175242 Free PMC article. - p14ARF activates a Tip60-dependent and p53-independent ATM/ATR/CHK pathway in response to genotoxic stress.
Eymin B, Claverie P, Salon C, Leduc C, Col E, Brambilla E, Khochbin S, Gazzeri S. Eymin B, et al. Mol Cell Biol. 2006 Jun;26(11):4339-50. doi: 10.1128/MCB.02240-05. Mol Cell Biol. 2006. PMID: 16705183 Free PMC article. - E2F3-a novel repressor of the ARF/p53 pathway.
Ginsberg D. Ginsberg D. Dev Cell. 2004 Jun;6(6):742-3. doi: 10.1016/j.devcel.2004.05.012. Dev Cell. 2004. PMID: 15177020 Review. - The dual specificity phosphatase Cdc25C is a direct target for transcriptional repression by the tumor suppressor p53.
St Clair S, Manfredi JJ. St Clair S, et al. Cell Cycle. 2006 Apr;5(7):709-13. doi: 10.4161/cc.5.7.2628. Epub 2006 Apr 1. Cell Cycle. 2006. PMID: 16582636 Review.
Cited by
- YEATS4 is a novel oncogene amplified in non-small cell lung cancer that regulates the p53 pathway.
Pikor LA, Lockwood WW, Thu KL, Vucic EA, Chari R, Gazdar AF, Lam S, Lam WL. Pikor LA, et al. Cancer Res. 2013 Dec 15;73(24):7301-12. doi: 10.1158/0008-5472.CAN-13-1897. Epub 2013 Oct 29. Cancer Res. 2013. PMID: 24170126 Free PMC article. - Evolutionary conserved relocation of chromatin remodeling complexes to the mitotic apparatus.
Messina G, Prozzillo Y, Monache FD, Santopietro MV, Dimitri P. Messina G, et al. BMC Biol. 2022 Aug 3;20(1):172. doi: 10.1186/s12915-022-01365-5. BMC Biol. 2022. PMID: 35922843 Free PMC article. - Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation.
Wang Y, Jin J, Chung MWH, Feng L, Sun H, Hao Q. Wang Y, et al. Proc Natl Acad Sci U S A. 2018 Mar 6;115(10):2365-2370. doi: 10.1073/pnas.1717664115. Epub 2018 Feb 20. Proc Natl Acad Sci U S A. 2018. PMID: 29463709 Free PMC article. - Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer.
Hsu CC, Shi J, Yuan C, Zhao D, Jiang S, Lyu J, Wang X, Li H, Wen H, Li W, Shi X. Hsu CC, et al. Genes Dev. 2018 Jan 1;32(1):58-69. doi: 10.1101/gad.303784.117. Epub 2018 Feb 1. Genes Dev. 2018. PMID: 29437725 Free PMC article. - Targeting the p53-MDM2 interaction by the small-molecule MDM2 antagonist Nutlin-3a: a new challenged target therapy in adult Philadelphia positive acute lymphoblastic leukemia patients.
Trino S, Iacobucci I, Erriquez D, Laurenzana I, De Luca L, Ferrari A, Ghelli Luserna Di Rorà A, Papayannidis C, Derenzini E, Simonetti G, Lonetti A, Venturi C, Cattina F, Ottaviani E, Abbenante MC, Russo D, Perini G, Musto P, Martinelli G. Trino S, et al. Oncotarget. 2016 Mar 15;7(11):12951-61. doi: 10.18632/oncotarget.7339. Oncotarget. 2016. PMID: 26887044 Free PMC article.
References
- An, W., and R. G. Roeder. 2004. Reconstitution and transcriptional analysis of chromatin in vitro. Methods Enzymol. 377:460-474. - PubMed
- Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous