Central role for MyD88 in the responses of microglia to pathogen-associated molecular patterns - PubMed (original) (raw)
Central role for MyD88 in the responses of microglia to pathogen-associated molecular patterns
Nilufer Esen et al. J Immunol. 2006.
Abstract
Microglia, the innate immune effector cells of the CNS parenchyma, express TLR that recognize conserved motifs of microorganisms referred to as pathogen-associated molecular patterns (PAMP). All TLRs identified to date, with the exception of TLR3, use a common adaptor protein, MyD88, to transduce activation signals. Recently, we reported that microglial activation in response to the Gram-positive bacterium Staphylococcus aureus was not completely attenuated following TLR2 ablation, suggesting the involvement of additional receptors. To assess the functional role of alternative TLRs in microglial responses to S. aureus and its cell wall product peptidoglycan as well as the Gram-negative PAMP LPS, we evaluated primary microglia from MyD88 knockout (KO) and wild-type mice. The induction of TNF-alpha, IL-12 p40, and MIP-2 (CXCL2) expression by S. aureus- and peptidoglycan-stimulated microglia was MyD88 dependent, as revealed by the complete inhibition of cytokine production in MyD88 KO cells. In addition, the expression of additional pattern recognition receptors, including TLR9, pentraxin-3, and lectin-like oxidized LDL receptor-1, was regulated, in part, via a MyD88-dependent manner as demonstrated by the attenuated expression of these receptors in MyD88 KO microglia. Microglial activation was only partially inhibited in LPS-stimulated MyD88 KO cells, suggesting the involvement of MyD88-independent pathways. Collectively, these findings reveal the complex mechanisms for microglia to respond to diverse bacterial pathogens, which occur via both MyD88-dependent and -independent pathways.
Conflict of interest statement
Disclosures The authors have no financial conflict of interest.
Figures
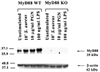
FIGURE 1
MyD88 is induced in primary microglia in response to both Gram-positive and -negative bacterial stimuli. MyD88 KO and WT primary microglia were stimulated with either 107 heat-inactivated S. aureus, 10µg/ml PGN, or 100 ng/ml LPS for 24 h, whereupon protein extracts from whole cell lysates (40 µg/sample) were evaluated for MyD88 expression by Western blotting as described in Materials and Methods. Following analysis, blots were stripped and reprobed with an Ab specific for β-actin to verify uniformity in gel loading. Results are representative of three independent experiments.
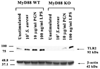
FIGURE 2
LPS-induced TLR2 expression is regulated by a MyD88- independent pathway. MyD88 KO and WT primary microglia were stimulated with either 107 heat-inactivated S. aureus, 10 µg/ml PGN, or 100 ng/ml LPS for 24 h, whereupon protein extracts from whole cell lysates (40 µg/sample) were evaluated for TLR2 expression by Western blotting as described in Materials and Methods. Following analysis, blots were stripped and reprobed with an Ab specific for β-actin to verify uniformity in gel loading. Results are representative of three independent experiments.
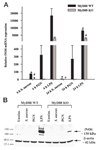
FIGURE 3
Role of MyD88-dependent signals in regulating iNOS expression. A, MyD88 KO and WT microglia were seeded at 2 × 106 cells/ well in 6-well plates and incubated overnight. The following day, cells were stimulated with 107 heat-inactivated S. aureus, 10 µg/ml PGN, or 100 ng/ml LPS for 6 or 24 h, whereupon total RNA was isolated and examined for iNOS expression by qRT-PCR as described in Materials and Methods. Gene expression levels were calculated after normalizing iNOS signals against GAPDH and are presented in relative mRNA expression units (mean + SEM of three independent experiments). Significant differences in iNOS mRNA expression between unstimulated (data not shown) vs PAMP-treated microglia are denoted with asterisks (*, p < 0.05), whereas significant differences between MyD88 KO vs WT microglia are denoted with a hatched sign (#, p < 0.05). B, MyD88 KO and WT primary microglia were stimulated with either 107 heat-inactivated S. aureus, 10 µg/ml PGN, or 100 ng/ml LPS for 24 h, whereupon protein extracts from whole cell lysates (40 µg/sample) were evaluated for iNOS expression by Western blotting as described in Materials and Methods. Following analysis, blots were stripped and reprobed with an Ab specific for β-actin to verify uniformity in gel loading. Results are representative of three independent experiments.
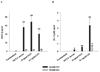
FIGURE 4
MyD88 is essential in mediating microglial activation in response to S. aureus and PGN. MyD88 KO and WT microglia were seeded at 2 × 105 cells/well in 96-well plates and incubated overnight. The following day, cells were exposed to various concentrations of heat-inactivated S. aureus (A), PGN (B), or LPS (C) for 24 h, whereupon conditioned supernatants were collected and analyzed for TNF-α protein expression by ELISA. Results are presented as the amount of TNF-α(nanogram) per milliliter of culture supernatant (mean ± SD). Microglial cell viability was assessed using a standard MTT assay and the raw OD570 absorbance values are reported (D; mean ± SD). Significant differences in TNF-α expression between unstimulated vs PAMP-treated microglia are denoted with asterisks (*, p < 0.05; **, p < 0.001), whereas significant differences between MyD88 KO vs WT microglia are denoted with a hatched sign (##, p < 0.001). Results are representative of three independent experiments.
FIGURE 5
The loss of MyD88 results in diminished MIP-2 and IL-12 p40 expression. Primary microglia from MyD88 KO and WT mice were seeded at 2 × 105 cells/well in 96-well plates and incubated overnight. The following day, cells were exposed to either 107 heat-inactivated S. aureus, 10 µg/ml PGN, or 100 ng/ml LPS for 24 h, whereupon conditioned supernatants were collected and analyzed for MIP-2 (A) and IL-12 p40 (B) protein expression by ELISA. Results are presented as the amount of cytokine (nanograms) per milliliter of culture supernatant (mean ± SD). Significant differences in cytokine expression between unstimulated vs PAMP-treated microglia are denoted with asterisks (*, p < 0.05; **, p < 0.001), whereas significant differences between MyD88 KO vs WT micro-glia are denoted with a hatched sign (##, p < 0.001). Results are representative of three independent experiments.
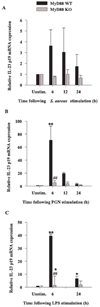
FIGURE 6
MyD88 is involved in IL-23 p19 mRNA expression. MyD88 KO and WT microglia were seeded at 2 × 106 cells/well in 6-well plates and incubated overnight. The following day, cells were stimulated with either 107 heat-inactivated S. aureus (A), 10 µg/ml PGN (B), or 100 ng/ml LPS (C) for 6, 12, or 24 h, whereupon total RNA was isolated and examined for IL-23 p19 expression by qRT-PCR as described in Materials and Methods. Gene expression levels were calculated after normalizing IL-23 p19 signals against GAPDH and are presented in relative mRNA expression units (mean ± SEM of three independent experiments). Significant differences in IL-23 p19 mRNA expression between unstimulated vs PAMP-treated microglia are denoted with asterisks (*, p < 0.05; **, p < 0.001), whereas significant differences between MyD88 KO vs WT microglia are denoted with a hatched sign (##, p < 0.001).
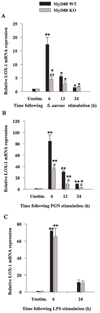
FIGURE 7
Microglial LOX-1 expression is partially regulated by MyD88. MyD88 KO and WT microglia were seeded at 2 × 106 cells/well in 6-well plates and incubated overnight. The following day, cells were stimulated with either 107 heat-inactivated S. aureus (A), 10 µg/ml PGN (B), or 100 ng/ml LPS (C) for 6, 12, or 24 h, whereupon total RNA was isolated and examined for LOX-1 expression by qRT-PCR as described in Materials and Methods. Gene expression levels were calculated after normalizing LOX-1 signals against GAPDH and are presented in relative mRNA expression units (mean ± SEM of three independent experiments). Significant differences in LOX-1 mRNA expression between unstimulated vs PAMP-treated microglia are denoted with asterisks (*, p < 0.05; **, p < 0.001), whereas significant differences between MyD88 KO vs WT microglia are denoted with a hatched sign (#, p < 0.05; ##, p < 0.001).
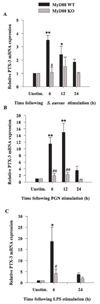
FIGURE 8
The expression of PTX3 is regulated by MyD88 in primary microglia. MyD88 KO and WT microglia were seeded at 2 × 106 cells/ well in 6-well plates and incubated overnight. The following day, cells were stimulated with either 107 heat-inactivated S. aureus (A), 10 µg/ml PGN (B), or 100 ng/ml LPS (C) for 6, 12, or 24 h, whereupon total RNA was isolated and examined for PTX3 expression by qRT-PCR as described in Materials and Methods. Gene expression levels were calculated after normalizing PTX3 signals against GAPDH and are presented in relative mRNA expression units (mean ± SEM of three independent experiments). Significant differences in PTX3 mRNA expression between unstimulated vs PAMP-treated microglia are denoted with asterisks (*, p < 0.05; **, p < 0.001), whereas significant differences between MyD88 KO vs WT microglia are denoted with a hatched sign (#, p < 0.05; ##, p < 0.001).
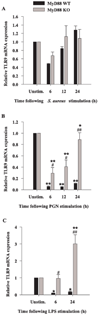
FIGURE 9
The loss of MyD88 results in differential expression of TLR9 mRNA expression in response to S. aureus, PGN, or LPS. MyD88 KO and WT microglia were seeded at 2 × 106 cells/well in 6-well plates and incubated overnight. The following day, cells were stimulated with either 107 heat-inactivated S. aureus (A), 10 µg/ml PGN (B), or 100 ng/ml LPS (C) for 6, 12, or 24 h, whereupon total RNA was isolated and examined for TLR9 expression by qRT-PCR as described in Materials and Methods. Gene expression levels were calculated after normalizing TLR9 signals against GAPDH and are presented in relative mRNA expression units (mean ± SEM of three independent experiments). Significant differences in TLR9 mRNA expression between unstimulated vs PAMP-treated microglia are denoted with asterisks (*, p < 0.05; **, p < 0.001), whereas significant differences between MyD88 KO vs WT microglia are denoted with a hatched sign (#, p < 0.05; ##, p < 0.001).
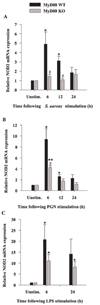
FIGURE 10
NOD2 mRNA is expressed in primary microglia. MyD88 KO and WT microglia were seeded at 2 × 106 cells/well in 6-well plates and incubated overnight. The following day, cells were stimulated with either 107 heat-inactivated S. aureus (A), 10 µg/ml PGN (B), or 100 ng/ml LPS (C) for 6, 12, or 24 h, whereupon total RNA was isolated and examined for NOD2 expression by qRT-PCR as described in Materials and Methods. Gene expression levels were calculated after normalizing NOD2 signals against GADPH and are presented in relative mRNA expression units (mean ± SEM of three independent experiments). Significant differences in NOD2 mRNA expression between unstimulated vs PAMP-treated microglia are denoted with asterisks (*, p < 0.05; **, p < 0.001), whereas significant differences between MyD88 KO vs WT microglia are denoted with a hatched sign (#, p < 0.05).
Similar articles
- Toll-like receptor 2 (TLR2) is pivotal for recognition of S. aureus peptidoglycan but not intact bacteria by microglia.
Kielian T, Esen N, Bearden ED. Kielian T, et al. Glia. 2005 Mar;49(4):567-76. doi: 10.1002/glia.20144. Glia. 2005. PMID: 15593098 Free PMC article. - Effects of low dose GM-CSF on microglial inflammatory profiles to diverse pathogen-associated molecular patterns (PAMPs).
Esen N, Kielian T. Esen N, et al. J Neuroinflammation. 2007 Mar 20;4:10. doi: 10.1186/1742-2094-4-10. J Neuroinflammation. 2007. PMID: 17374157 Free PMC article. - Recognition of Staphylococcus aureus-derived peptidoglycan (PGN) but not intact bacteria is mediated by CD14 in microglia.
Esen N, Kielian T. Esen N, et al. J Neuroimmunol. 2005 Dec 30;170(1-2):93-104. doi: 10.1016/j.jneuroim.2005.09.003. Epub 2005 Oct 17. J Neuroimmunol. 2005. PMID: 16229899 Free PMC article. - Microglial activation by Citrobacter koseri is mediated by TLR4- and MyD88-dependent pathways.
Liu S, Kielian T. Liu S, et al. J Immunol. 2009 Nov 1;183(9):5537-47. doi: 10.4049/jimmunol.0900083. Epub 2009 Oct 7. J Immunol. 2009. PMID: 19812209 Free PMC article. - Microglia in infectious diseases of the central nervous system.
Mariani MM, Kielian T. Mariani MM, et al. J Neuroimmune Pharmacol. 2009 Dec;4(4):448-61. doi: 10.1007/s11481-009-9170-6. Epub 2009 Sep 2. J Neuroimmune Pharmacol. 2009. PMID: 19728102 Free PMC article. Review.
Cited by
- Toll-like receptors in brain abscess.
Esen N, Kielian T. Esen N, et al. Curr Top Microbiol Immunol. 2009;336:41-61. doi: 10.1007/978-3-642-00549-7_3. Curr Top Microbiol Immunol. 2009. PMID: 19688327 Free PMC article. Review. - Anti-inflammatory efficacy of dexamethasone and Nrf2 activators in the CNS using brain slices as a model of acute injury.
Graber DJ, Hickey WF, Stommel EW, Harris BT. Graber DJ, et al. J Neuroimmune Pharmacol. 2012 Mar;7(1):266-78. doi: 10.1007/s11481-011-9338-8. Epub 2012 Jan 17. J Neuroimmune Pharmacol. 2012. PMID: 22249489 - Exosomes derived from mycobacterium tuberculosis-infected MSCs induce a pro-inflammatory response of macrophages.
Liu M, Wang Z, Ren S, Zhao H. Liu M, et al. Aging (Albany NY). 2021 Apr 19;13(8):11595-11609. doi: 10.18632/aging.202854. Epub 2021 Apr 19. Aging (Albany NY). 2021. PMID: 33872217 Free PMC article. - HMGB1, an innate alarmin, in the pathogenesis of type 1 diabetes.
Zhang S, Zhong J, Yang P, Gong F, Wang CY. Zhang S, et al. Int J Clin Exp Pathol. 2009 Sep 8;3(1):24-38. Int J Clin Exp Pathol. 2009. PMID: 19918326 Free PMC article. Review. - Cytokines and chemokines at the crossroads of neuroinflammation, neurodegeneration, and neuropathic pain.
Ramesh G, MacLean AG, Philipp MT. Ramesh G, et al. Mediators Inflamm. 2013;2013:480739. doi: 10.1155/2013/480739. Epub 2013 Aug 12. Mediators Inflamm. 2013. PMID: 23997430 Free PMC article. Review.
References
- Aloisi F. Immune function of microglia. Glia. 2001;36:165–179. - PubMed
- Kaisho T, Akira S. Pleiotropic function of Toll-like receptors. Microbes Infect. 2004;6:1388–1394. - PubMed
- Kopp E, Medzhitov R. Recognition of microbial infection by Toll-like receptors. Curr. Opin. Immunol. 2003;15:396–401. - PubMed
- Takeda K, Akira S. TLR signaling pathways. Semin. Immunol. 2004;16:3–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials