Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus - PubMed (original) (raw)
Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus
Paul R Gilson et al. Proc Natl Acad Sci U S A. 2006.
Abstract
The introduction of plastids into different heterotrophic protists created lineages of algae that diversified explosively, proliferated in marine and freshwater environments, and radically altered the biosphere. The origins of these secondary plastids are usually inferred from the presence of additional plastid membranes. However, two examples provide unique snapshots of secondary-endosymbiosis-in-action, because they retain a vestige of the endosymbiont nucleus known as the nucleomorph. These are chlorarachniophytes and cryptomonads, which acquired their plastids from a green and red alga respectively. To allow comparisons between them, we have sequenced the nucleomorph genome from the chlorarachniophyte Bigelowiella natans: at a mere 373,000 bp and with only 331 genes, the smallest nuclear genome known and a model for extreme reduction. The genome is eukaryotic in nature, with three linear chromosomes containing densely packed genes with numerous overlaps. The genome is replete with 852 introns, but these are the smallest introns known, being only 18, 19, 20, or 21 nt in length. These pygmy introns are shown to be miniaturized versions of normal-sized introns present in the endosymbiont at the time of capture. Seventeen nucleomorph genes encode proteins that function in the plastid. The other nucleomorph genes are housekeeping entities, presumably underpinning maintenance and expression of these plastid proteins. Chlorarachniophyte plastids are thus serviced by three different genomes (plastid, nucleomorph, and host nucleus) requiring remarkable coordination and targeting. Although originating by two independent endosymbioses, chlorarachniophyte and cryptomonad nucleomorph genomes have converged upon remarkably similar architectures but differ in many molecular details that reflect two distinct trajectories to hypercompaction and reduction.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
Evolution of chlorarachniophytes such as B. natans by sequential endosymbioses. Enslavement of a photosynthetic, cyanobacterium-like prokaryote (Cb) introduces photosynthesis into a eukaryotic host (Euk 1), whose nucleus (Nu1) acquires at least 1,000 cyanobacterial genes over time. Secondary endosymbiosis involves capture and retention of the primary photosynthetic eukaryote by another eukaryote (Euk 2), producing a plastid with four bounding membranes such as those of cryptomonads, chlorarachniophytes, haptophyte and heterokont algae, and malaria parasites. Essential plastid protein genes are transferred from the endosymbiont nucleus (Nm, nucleomorph) to the nucleus (Nu2) of the second eukaryotic host. Here, we show that only 17 of the original plastid protein genes remain in the nucleomorph of B. natans, preventing its loss.
Fig. 2.
Gene map of the three B. natans nucleomorph chromosomes. Inverted terminal repeats are shaded in yellow. Pygmy introns are depicted as triangles. Genes for which excision of the pygmy introns (where present) has been confirmed by cDNA analysis are named in red type.
Fig. 3.
Nucleomorph genes harbor numerous ultra-small introns. (A) Consensus plots of the 18-, 19-, 20- and 21-nt introns showing 5′-GT. . .AG-3′ borders but no exon or intron core consensus other than an A at −2. This A and overall high AT content are the only discernible differences between real introns and 19-nt exonic sequences with 5′-GT. . .AG-3′ borders (pseudointrons) that are not removed by the spliceosome. (B) Nineteen nucleotide introns are the most abundant in the nucleomorph, but equivalent numbers of each size category of pseudointrons occur. (C) Numbers of introns shared between the nucleomorph, Arabidopsis, and Chlamydomonas. Most nucleomorph introns occur at the same position as large introns in these closely related genomes, suggesting that the pygmy introns began as normal introns but have been reduced by DNA loss to converge on a minimal spliceable size, ≈19 nt.
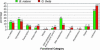
Fig. 4.
Comparison of gene functional categories from the nucleomorph genomes of B. natans and G. theta. Percentages of genes belonging to different functional categories are indicated by the scale bar on the left, and the total numbers of genes belonging to these categories are shown above the class bars.
Comment in
- The tiny enslaved genome of a rhizarian alga.
Cavalier-Smith T. Cavalier-Smith T. Proc Natl Acad Sci U S A. 2006 Jun 20;103(25):9379-80. doi: 10.1073/pnas.0603505103. Epub 2006 Jun 12. Proc Natl Acad Sci U S A. 2006. PMID: 16769883 Free PMC article. No abstract available.
Similar articles
- Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: convergent reductive evolution and frequent recombination in nucleomorph-bearing algae.
Tanifuji G, Onodera NT, Brown MW, Curtis BA, Roger AJ, Ka-Shu Wong G, Melkonian M, Archibald JM. Tanifuji G, et al. BMC Genomics. 2014 May 15;15(1):374. doi: 10.1186/1471-2164-15-374. BMC Genomics. 2014. PMID: 24885563 Free PMC article. - Nucleomorph Genome Sequences of Two Chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata.
Suzuki S, Shirato S, Hirakawa Y, Ishida K. Suzuki S, et al. Genome Biol Evol. 2015 May 22;7(6):1533-45. doi: 10.1093/gbe/evv096. Genome Biol Evol. 2015. PMID: 26002880 Free PMC article. - Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set.
Tanifuji G, Onodera NT, Wheeler TJ, Dlutek M, Donaher N, Archibald JM. Tanifuji G, et al. Genome Biol Evol. 2011;3:44-54. doi: 10.1093/gbe/evq082. Epub 2010 Dec 8. Genome Biol Evol. 2011. PMID: 21147880 Free PMC article. - Good things in small packages: the tiny genomes of chlorarachniophyte endosymbionts.
Gilson PR, McFadden GI. Gilson PR, et al. Bioessays. 1997 Feb;19(2):167-73. doi: 10.1002/bies.950190212. Bioessays. 1997. PMID: 9046247 Review. - Going, going, not quite gone: nucleomorphs as a case study in nuclear genome reduction.
Archibald JM, Lane CE. Archibald JM, et al. J Hered. 2009 Sep-Oct;100(5):582-90. doi: 10.1093/jhered/esp055. Epub 2009 Jul 17. J Hered. 2009. PMID: 19617523 Review.
Cited by
- The complete mitochondrial genome assembly of Capsicum pubescens reveals key evolutionary characteristics of mitochondrial genes of two Capsicum subspecies.
Li L, Fu H, Altaf MA, Wang Z, Lu X. Li L, et al. BMC Genomics. 2024 Nov 11;25(1):1064. doi: 10.1186/s12864-024-10985-w. BMC Genomics. 2024. PMID: 39528932 Free PMC article. - Decryption of the survival "black box": gene family expansion promotes the encystment in ciliated protists.
Jin D, Li C, Chen X, Wang Y, Al-Rasheid KAS, Stover NA, Shao C, Zhang T. Jin D, et al. BMC Genomics. 2024 Mar 18;25(1):286. doi: 10.1186/s12864-024-10207-3. BMC Genomics. 2024. PMID: 38500030 Free PMC article. - Discovery of a Novel Intron in US10/US11/US12 of HSV-1 Strain 17.
Chang W, Hao M, Qiu J, Sherman BT, Imamichi T. Chang W, et al. Viruses. 2023 Oct 25;15(11):2144. doi: 10.3390/v15112144. Viruses. 2023. PMID: 38005822 Free PMC article. - Contrasting outcomes of genome reduction in mikrocytids and microsporidians.
Žárský V, Karnkowska A, Boscaro V, Trznadel M, Whelan TA, Hiltunen-Thorén M, Onut-Brännström I, Abbott CL, Fast NM, Burki F, Keeling PJ. Žárský V, et al. BMC Biol. 2023 Jun 6;21(1):137. doi: 10.1186/s12915-023-01635-w. BMC Biol. 2023. PMID: 37280585 Free PMC article. - Gene loss, pseudogenization, and independent genome reduction in non-photosynthetic species of Cryptomonas (Cryptophyceae) revealed by comparative nucleomorph genomics.
Kim JI, Tanifuji G, Jeong M, Shin W, Archibald JM. Kim JI, et al. BMC Biol. 2022 Oct 8;20(1):227. doi: 10.1186/s12915-022-01429-6. BMC Biol. 2022. PMID: 36209116 Free PMC article.
References
- Archibald J. M. IUBMB Life. 2005;57:539–547. - PubMed
- Cavalier-Smith T., Lee J. J. Protozool. 1985;32:376–379.
- McFadden G. I. J. Phycol. 2001;37:1–9.
- Soll J., Schleiff E. Nat. Rev. Mol. Cell Biol. 2004;5:198–208. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials