Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase - PubMed (original) (raw)
Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase
Stephen E Moyer et al. Proc Natl Acad Sci U S A. 2006.
Abstract
The protein Cdc45 plays a critical but poorly understood role in the initiation and elongation stages of eukaryotic DNA replication. To study Cdc45's function in DNA replication, we purified Cdc45 protein from Drosophila embryo extracts by a combination of traditional and immunoaffinity chromatography steps and found that the protein exists in a stable, high-molecular-weight complex with the Mcm2-7 hexamer and the GINS tetramer. The purified Cdc45/Mcm2-7/GINS complex is associated with an active ATP-dependent DNA helicase function. RNA interference knock-down experiments targeting the GINS and Cdc45 components establish that the proteins are required for the S phase transition in Drosophila cells. The data suggest that this complex forms the core helicase machinery for eukaryotic DNA replication.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
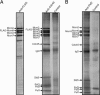
Purification of the CMG complex. (A) Purification schematic. See the supporting information for additional details. (B) Material eluted from the anti-Cdc45 IP after the Mono Q step was separated by SDS/PAGE and visualized by Deep Purple stain. For improved resolution of individual bands, the image is composed of the eluate separated by two different acrylamide gels: 9% (top of gel to IgG) and 12% (IgG to bottom of gel). The complete lane of each gel used for this image is shown in Fig. 6. All subsequent gels in this study are 10% acrylamide and contain all proteins in one complete lane. (C) At the Superdex-200 step shown in A, each fraction was subjected to an anti-Cdc45 IP, and the individual eluates were separated on SDS/PAGE and immunoblotted with antibodies against the indicated proteins. The antibodies have variable staining intensity; the ratio of all CMG proteins to each other in the high-molecular-weight fractions is the same (see Figs. 1_B_, 2, and 6).
Fig. 2.
IPs with antibodies against different CMG complex members. In this figure, the proteins were identified by immunoblot and _R_f value. (A) (Left) FLAG-Mcm6 material purified from embryo extracts by anti-FLAG chromatography. (Right) From the material first purified by the anti-FLAG chromatography, an anti-Cdc45 IP and a control (rabbit IgG) IP were performed. The anti-Cdc45 immunoprecipitated material shown here is enriched for Cdc45 ≈70-fold over the purified FLAG-Mcm6 material (i.e., ≈700 μl of the material in Left was used to obtain 10 μl of the material in Right). Both gels were stained with Deep Purple. (B) Peak Cdc45-containing fractions from the Mono Q step were precipitated with anti-Psf2 antibodies and control antibodies (anti-FLAG), and the eluates were stained with Deep Purple.
Fig. 3.
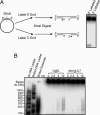
The CMG complex is a helicase. (A) Schematic of the purification protocol used for the helicase assay. (B) Illustration of helicase substrate (not to scale). (C) (Top) PhosphorImager picture of helicase assays with anti-Cdc45 peptide B-purified material from each Mono Q fraction. (Middle) Quantitation of the percentage of primers displaced from duplex DNA in each helicase reaction. (Bottom) Immunoblot with antibodies against indicated proteins of the anti-Cdc45 peptide B-purified material from each Mono Q fraction. The Cdc45 protein peak in fractions 12–15 is monomeric Cdc45 that has copurified with the CMG complex until this chromatography step. (Left) Deep Purple stain of anti-Cdc45 peptide B-purified material from fraction 20. (D) (Left) Immunoblots of material that has been immunodepleted with anti-Psf2, anti-Mcm5, or control antibodies. (Right) PhosphorImager picture of helicase assays with protein purified by anti-Cdc45 peptide B IPs from material previously depleted with anti-Psf2, anti-Mcm5, or control antibodies. (E) (Left) Helicase assays with material mock-eluted or Cdc45 peptide B-eluted from anti-Cdc45 peptide B IPs from purified FLAG-Mcm6 material. (Center) Helicase assays with and without ATP with CMG complex isolated from purified FLAG-Mcm6 material. (Right) Helicase assays with CMG complex purified as in A and with varying concentrations of ATP.
Fig. 4.
CMG directionality and processivity. (A) (Left) Cartoon of procedure for preparing substrates for directionality assays. A 48-mer primer was annealed to m13mp18 ssDNA plasmid and then labeled with 32P at the 5′ end with T4 polynucleotide kinase and [γ-32P]ATP or at the 3′ end with T7 DNA polymerase and [α-32P]dGTP. The duplex region was digested with SmaI, creating a linear plasmid with ssDNA in the middle and duplex DNA at both ends (the 5′ labeled primer is 20 bases, and the 3′ labeled primer is 28 bases). The radiolabeled primer can only be approached on ssDNA from one direction, thereby allowing determination of the directionality of helicase movement on DNA. (Right) PhosphorImager picture of CMG complex tested with both directionality substrates. (B) The primer in Fig. 3_B_ was extended by the DNA polymerase of T7 (Sequenase), and the heterogeneous products created substrates for helicase assays. A PhosphorImager picture of DNA substrates after incubation in helicase assays with increasing amounts of CMG or Mcm4,6,7 complexes is shown. For measuring the lengths of the duplex DNA in the substrates, a dsDNA ladder was labeled with 32P and separated on the gel in either native or boiled form.
Fig. 5.
Cdc45 and GINS proteins are required for normal S phase progression. (A) (Left) Drosophila Kc tissue culture cells were treated with ≈500 bp of dsRNA directed against Cdc45, Sld5, Psf1, Psf2, Psf3, or nonspecific (NS) RNA. After 8 days of RNAi treatment, cells were harvested; FACS profiles are shown. The x axis is arbitrary fluorescence units, and the y axis is the number of cells (0–128). (Right) Cell cycle distribution of RNAi-treated cells. (B) A model of the CMG complex. Green, Mcm2–7; blue, GINS; yellow, Cdc45.
Similar articles
- Dpb11 protein helps control assembly of the Cdc45·Mcm2-7·GINS replication fork helicase.
Dhingra N, Bruck I, Smith S, Ning B, Kaplan DL. Dhingra N, et al. J Biol Chem. 2015 Mar 20;290(12):7586-601. doi: 10.1074/jbc.M115.640383. Epub 2015 Feb 6. J Biol Chem. 2015. PMID: 25659432 Free PMC article. - Mcm10 coordinates the timely assembly and activation of the replication fork helicase.
Perez-Arnaiz P, Bruck I, Kaplan DL. Perez-Arnaiz P, et al. Nucleic Acids Res. 2016 Jan 8;44(1):315-29. doi: 10.1093/nar/gkv1260. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26582917 Free PMC article. - Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins.
Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Ilves I, et al. Mol Cell. 2010 Jan 29;37(2):247-58. doi: 10.1016/j.molcel.2009.12.030. Mol Cell. 2010. PMID: 20122406 Free PMC article. - The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism.
Li H, O'Donnell ME. Li H, et al. Bioessays. 2018 Mar;40(3):10.1002/bies.201700208. doi: 10.1002/bies.201700208. Epub 2018 Feb 6. Bioessays. 2018. PMID: 29405332 Free PMC article. Review. - The eukaryotic Mcm2-7 replicative helicase.
Vijayraghavan S, Schwacha A. Vijayraghavan S, et al. Subcell Biochem. 2012;62:113-34. doi: 10.1007/978-94-007-4572-8_7. Subcell Biochem. 2012. PMID: 22918583 Review.
Cited by
- Regulating DNA replication in eukarya.
Siddiqui K, On KF, Diffley JF. Siddiqui K, et al. Cold Spring Harb Perspect Biol. 2013 Sep 1;5(9):a012930. doi: 10.1101/cshperspect.a012930. Cold Spring Harb Perspect Biol. 2013. PMID: 23838438 Free PMC article. Review. - DNA Interactions Probed by Hydrogen-Deuterium Exchange (HDX) Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Confirm External Binding Sites on the Minichromosomal Maintenance (MCM) Helicase.
Graham BW, Tao Y, Dodge KL, Thaxton CT, Olaso D, Young NL, Marshall AG, Trakselis MA. Graham BW, et al. J Biol Chem. 2016 Jun 10;291(24):12467-12480. doi: 10.1074/jbc.M116.719591. Epub 2016 Apr 4. J Biol Chem. 2016. PMID: 27044751 Free PMC article. - Structure of a dimer of the Sulfolobus solfataricus MCM N-terminal domain reveals a potential role in MCM ring opening.
Meagher M, Spence MN, Enemark EJ. Meagher M, et al. Acta Crystallogr F Struct Biol Commun. 2021 Jun 1;77(Pt 6):177-186. doi: 10.1107/S2053230X21005331. Epub 2021 Jun 1. Acta Crystallogr F Struct Biol Commun. 2021. PMID: 34100776 Free PMC article. - RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks.
Hashimoto Y, Puddu F, Costanzo V. Hashimoto Y, et al. Nat Struct Mol Biol. 2011 Dec 4;19(1):17-24. doi: 10.1038/nsmb.2177. Nat Struct Mol Biol. 2011. PMID: 22139015 Free PMC article. - Distinct epigenetic features of differentiation-regulated replication origins.
Smith OK, Kim R, Fu H, Martin MM, Lin CM, Utani K, Zhang Y, Marks AB, Lalande M, Chamberlain S, Libbrecht MW, Bouhassira EE, Ryan MC, Noble WS, Aladjem MI. Smith OK, et al. Epigenetics Chromatin. 2016 May 10;9:18. doi: 10.1186/s13072-016-0067-3. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27168766 Free PMC article.
References
- Takahashi T. S., Wigley D. B., Walter J. C. Trends Biochem. Sci. 2005;30:437–444. - PubMed
- Labib K., Tercero J. A., Diffley J. F. Science. 2000;288:1643–1647. - PubMed
- Shechter D., Ying C. Y., Gautier J. J. Biol. Chem. 2004;279:45586–45593. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous