The MPI Bioinformatics Toolkit for protein sequence analysis - PubMed (original) (raw)
The MPI Bioinformatics Toolkit for protein sequence analysis
Andreas Biegert et al. Nucleic Acids Res. 2006.
Abstract
The MPI Bioinformatics Toolkit is an interactive web service which offers access to a great variety of public and in-house bioinformatics tools. They are grouped into different sections that support sequence searches, multiple alignment, secondary and tertiary structure prediction and classification. Several public tools are offered in customized versions that extend their functionality. For example, PSI-BLAST can be run against regularly updated standard databases, customized user databases or selectable sets of genomes. Another tool, Quick2D, integrates the results of various secondary structure, transmembrane and disorder prediction programs into one view. The Toolkit provides a friendly and intuitive user interface with an online help facility. As a key feature, various tools are interconnected so that the results of one tool can be forwarded to other tools. One could run PSI-BLAST, parse out a multiple alignment of selected hits and send the results to a cluster analysis tool. The Toolkit framework and the tools developed in-house will be packaged and freely available under the GNU Lesser General Public Licence (LGPL). The Toolkit can be accessed at http://toolkit.tuebingen.mpg.de.
Figures
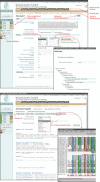
Figure 1
Input and result pages of PSI-BLAST with overlaid windows for genome databases and Jalview alignment viewer (32).
Similar articles
- Protein Sequence Analysis Using the MPI Bioinformatics Toolkit.
Gabler F, Nam SZ, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. Gabler F, et al. Curr Protoc Bioinformatics. 2020 Dec;72(1):e108. doi: 10.1002/cpbi.108. Curr Protoc Bioinformatics. 2020. PMID: 33315308 - The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis.
Alva V, Nam SZ, Söding J, Lupas AN. Alva V, et al. Nucleic Acids Res. 2016 Jul 8;44(W1):W410-5. doi: 10.1093/nar/gkw348. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131380 Free PMC article. - Windows .NET Network Distributed Basic Local Alignment Search Toolkit (W.ND-BLAST).
Dowd SE, Zaragoza J, Rodriguez JR, Oliver MJ, Payton PR. Dowd SE, et al. BMC Bioinformatics. 2005 Apr 8;6:93. doi: 10.1186/1471-2105-6-93. BMC Bioinformatics. 2005. PMID: 15819992 Free PMC article. - State-of-the-art bioinformatics protein structure prediction tools (Review).
Pavlopoulou A, Michalopoulos I. Pavlopoulou A, et al. Int J Mol Med. 2011 Sep;28(3):295-310. doi: 10.3892/ijmm.2011.705. Epub 2011 May 23. Int J Mol Med. 2011. PMID: 21617841 Review. - The BioTools Suite. A comprehensive suite of platform-independent bioinformatics tools.
Wishart DS, Fortin S. Wishart DS, et al. Mol Biotechnol. 2001 Sep;19(1):59-77. doi: 10.1385/MB:19:1:059. Mol Biotechnol. 2001. PMID: 11697221 Review.
Cited by
- Cystinosin, MPDU1, SWEETs and KDELR belong to a well-defined protein family with putative function of cargo receptors involved in vesicle trafficking.
Saudek V. Saudek V. PLoS One. 2012;7(2):e30876. doi: 10.1371/journal.pone.0030876. Epub 2012 Feb 17. PLoS One. 2012. PMID: 22363504 Free PMC article. - The S. pombe mRNA decapping complex recruits cofactors and an Edc1-like activator through a single dynamic surface.
Wurm JP, Overbeck J, Sprangers R. Wurm JP, et al. RNA. 2016 Sep;22(9):1360-72. doi: 10.1261/rna.057315.116. Epub 2016 Jun 28. RNA. 2016. PMID: 27354705 Free PMC article. - Structural conservation of the myoviridae phage tail sheath protein fold.
Aksyuk AA, Kurochkina LP, Fokine A, Forouhar F, Mesyanzhinov VV, Tong L, Rossmann MG. Aksyuk AA, et al. Structure. 2011 Dec 7;19(12):1885-94. doi: 10.1016/j.str.2011.09.012. Structure. 2011. PMID: 22153511 Free PMC article. - Crystal structure of the magnetobacterial protein MtxA C-terminal domain reveals a new sequence-structure relationship.
Davidov G, Müller FD, Baumgartner J, Bitton R, Faivre D, Schüler D, Zarivach R. Davidov G, et al. Front Mol Biosci. 2015 May 21;2:25. doi: 10.3389/fmolb.2015.00025. eCollection 2015. Front Mol Biosci. 2015. PMID: 26052516 Free PMC article. - Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals.
Shahid SA, Bardiaux B, Franks WT, Krabben L, Habeck M, van Rossum BJ, Linke D. Shahid SA, et al. Nat Methods. 2012 Dec;9(12):1212-7. doi: 10.1038/nmeth.2248. Epub 2012 Nov 11. Nat Methods. 2012. PMID: 23142870
References
- Subramaniam S. The biology workbench—a seamless database and analysis environment for the biologist. Proteins. 1998;32:1–2. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials