SLiMDisc: short, linear motif discovery, correcting for common evolutionary descent - PubMed (original) (raw)
SLiMDisc: short, linear motif discovery, correcting for common evolutionary descent
Norman E Davey et al. Nucleic Acids Res. 2006.
Abstract
Many important interactions of proteins are facilitated by short, linear motifs (SLiMs) within a protein's primary sequence. Our aim was to establish robust methods for discovering putative functional motifs. The strongest evidence for such motifs is obtained when the same motifs occur in unrelated proteins, evolving by convergence. In practise, searches for such motifs are often swamped by motifs shared in related proteins that are identical by descent. Prediction of motifs among sets of biologically related proteins, including those both with and without detectable similarity, were made using the TEIRESIAS algorithm. The number of motif occurrences arising through common evolutionary descent were normalized based on treatment of BLAST local alignments. Motifs were ranked according to a score derived from the product of the normalized number of occurrences and the information content. The method was shown to significantly outperform methods that do not discount evolutionary relatedness, when applied to known SLiMs from a subset of the eukaryotic linear motif (ELM) database. An implementation of Multiple Spanning Tree weighting outperformed two other weighting schemes, in a variety of settings.
Figures
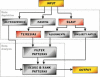
Figure 1
Simplified graphical representation of the SLiMDisc method. The steps completed by SLiMDisc are in green, those which occur outside the program are in red. The input dataset is given to the TEIRESIAS algorithm for pattern discovery and the BLAST algorithm to establish the evolutionary relationships of the parent proteins. The returned motifs are then filtered according to a number of user defined criteria. Finally, the motifs are ranked using information content (based on amino acid frequencies) and evolutionary relatedness.
Figure 2
Graphical representation of UHS and UP normalization techniques. Four proteins, labelled 1–4, are shown with annotated domains marked as coloured regions. Regions of homology as detected by BLAST are shown as grey boxes linking the sequences. Sequences 1 and 2 share a large homologous (orange) domain. Sequences 2 and 3 also share a homologous region but this is not annotated as a domain. Three other domains are specific to proteins 1 (green), 3 (blue) and 4 (purple). All motifs a–f have three occurrences in the dataset but have different support (shown in the table on the right) after filtering. a.→Motif a occurs in a shared region between 1 and 2, which is reduced by UHS to a single occurrence. The third occurrence in sequence 3 is not in an homologous region to 1 or 2 and is treated as a separate occurrence by UHS. However, proteins 2 and 3 share a homologous region and so UP will cluster sequences 1, 2 and 3, reducing the number of occurrences to 1. Filtering domains reduces the support to 1 in either case. b.→Motif b occurs in a shared region between 2 and 3, which is reduced by both UHS and UP to a single occurrence. This time, the third occurrence lies in the totally unrelated protein 4 and is counted with either filter. Filtering domains removed the occurrence in 4, reducing the support to 1. c.→Motif c lies purely within a repeated domain in protein 3. This is reduced to a single occurrence by both UHS and UP (the protein is homologous with itself). Although, whole-protein self-hits are ignored by UHS, the additional local BLAST hits between different domains (shown in grey) will still cause motif c to be filtered by UHS. Domain filtering removes it completely. d.→Motif d is the same as motif b, except that none of the occurrences lie in domains and so domain filtering makes no difference. e.→Motif e lies in non-homologous regions of protein 1 and 4. UHS therefore keeps all three occurrences. Whole-protein self-hits are ignored during the UHS filtering, and so both occurrences of motif e in protein 4 are counted. In contrast, UP clusters sequence 4 with itself and reduces the support to 2. No occurrences lie in domains and so domain filtering makes no difference. f.→Motif f is found in proteins 1, 3 and 4. None of these regions are homologous and so UHS gives a support of 3. UP, however, will group proteins 1 and 3; even though they do not directly share homology, they both share homology with common protein 2. UP therefore reduces the support to 2.
Figure 3
Scattergram of the information content versus the score for the KDEL (see Table 2) retrieving motif. Each blue point on the scattergram is a motif which has been considered by SLiMDisc. The points in green are the top three motifs ranked by the method. The actual SLiM for this dataset is the motif described by the regular expression [KRHQSAP][DENQT]EL.
Similar articles
- The SLiMDisc server: short, linear motif discovery in proteins.
Davey NE, Edwards RJ, Shields DC. Davey NE, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W455-9. doi: 10.1093/nar/gkm400. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576682 Free PMC article. - Masking residues using context-specific evolutionary conservation significantly improves short linear motif discovery.
Davey NE, Shields DC, Edwards RJ. Davey NE, et al. Bioinformatics. 2009 Feb 15;25(4):443-50. doi: 10.1093/bioinformatics/btn664. Epub 2009 Jan 9. Bioinformatics. 2009. PMID: 19136552 - DILIMOT: discovery of linear motifs in proteins.
Neduva V, Russell RB. Neduva V, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W350-5. doi: 10.1093/nar/gkl159. Nucleic Acids Res. 2006. PMID: 16845024 Free PMC article. - Discovering sequence motifs.
Bailey TL. Bailey TL. Methods Mol Biol. 2008;452:231-51. doi: 10.1007/978-1-60327-159-2_12. Methods Mol Biol. 2008. PMID: 18566768 Review. - Bioinformatics Approaches for Predicting Disordered Protein Motifs.
Bhowmick P, Guharoy M, Tompa P. Bhowmick P, et al. Adv Exp Med Biol. 2015;870:291-318. doi: 10.1007/978-3-319-20164-1_9. Adv Exp Med Biol. 2015. PMID: 26387106 Review.
Cited by
- Discovery and Characterization of Linear Motif Mediated Protein-Protein Complexes.
Zeke A, Alexa A, Reményi A. Zeke A, et al. Adv Exp Med Biol. 2024;3234:59-71. doi: 10.1007/978-3-031-52193-5_5. Adv Exp Med Biol. 2024. PMID: 38507200 - Whole-mitogenome analysis unveils previously undescribed genetic diversity in cane toads across their invasion trajectory.
Cheung K, Amos TG, Shine R, DeVore JL, Ducatez S, Edwards RJ, Rollins LA. Cheung K, et al. Ecol Evol. 2024 Mar 3;14(3):e11115. doi: 10.1002/ece3.11115. eCollection 2024 Mar. Ecol Evol. 2024. PMID: 38435005 Free PMC article. - The Australasian dingo archetype: de novo chromosome-length genome assembly, DNA methylome, and cranial morphology.
Ballard JWO, Field MA, Edwards RJ, Wilson LAB, Koungoulos LG, Rosen BD, Chernoff B, Dudchenko O, Omer A, Keilwagen J, Skvortsova K, Bogdanovic O, Chan E, Zammit R, Hayes V, Aiden EL. Ballard JWO, et al. Gigascience. 2023 Mar 20;12:giad018. doi: 10.1093/gigascience/giad018. Epub 2023 Mar 28. Gigascience. 2023. PMID: 36994871 Free PMC article. - Computational Prediction of Protein Intrinsically Disordered Region Related Interactions and Functions.
Han B, Ren C, Wang W, Li J, Gong X. Han B, et al. Genes (Basel). 2023 Feb 8;14(2):432. doi: 10.3390/genes14020432. Genes (Basel). 2023. PMID: 36833360 Free PMC article. Review. - The Australasian dingo archetype: De novo chromosome-length genome assembly, DNA methylome, and cranial morphology.
Ballard JWO, Field MA, Edwards RJ, Wilson LAB, Koungoulos LG, Rosen BD, Chernoff B, Dudchenko O, Omer A, Keilwagen J, Skvortsova K, Bogdanovic O, Chan E, Zammit R, Hayes V, Aiden EL. Ballard JWO, et al. bioRxiv [Preprint]. 2023 Jan 27:2023.01.26.525801. doi: 10.1101/2023.01.26.525801. bioRxiv. 2023. PMID: 36747621 Free PMC article. Updated. Preprint.
References
- Munro S., Pelham H.R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. - PubMed
- Furmanek A., Hofsteenge J. Protein C-mannosylation: facts and questions. Acta. Biochim. Pol. 2000;47:781–789. - PubMed
- Puntervoll P., Linding R., Gemund C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D.M., Ausiello G., Brannetti B., Costantini A., et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials