Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway - PubMed (original) (raw)
Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway
Malte Buchholz et al. EMBO J. 2006.
Abstract
The nuclear factor of activated T cell (NFAT) proteins are a family of Ca2+/calcineurin-responsive transcription factors primarily recognized for their central roles in T lymphocyte activation and cardiac valve development. We demonstrate that NFATc1 is commonly overexpressed in pancreatic carcinomas and enhances the malignant potential of tumor cells through transcriptional activation of the c-myc oncogene. Activated NFATc1 directly binds to a specific element within the proximal c-myc promoter and upregulates c-myc transcription, ultimately resulting in increased cell proliferation and enhanced anchorage-independent growth. Conversely, c-myc transcription and anchorage-dependent and -independent cell growth is significantly attenuated by inhibition of Ca2+/calcineurin signaling or siRNA-mediated knock down of NFATc1 expression. Together, these results demonstrate that ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway is an important mechanism of oncogenic c-myc activation in pancreatic cancer.
Figures
Figure 1
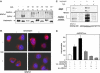
NFATc1 and calcineurin are ectopically expressed in pancreatic cancer. (A) Box-and-whisker plot illustrating normalized NFATc1 expression levels in pancreatic cancer (_n_=37) and normal/inflammatory control tissues (_n_=25) as determined in microarray experiments. (B) RT–PCR analyses using NFATc1-specific primers to validate the microarray data. Depicted are representative results for eight individual tissue samples. The human ribosomal protein, large, P0 gene (RPLP0) was used as housekeeping gene control. PaCa=pancreatic carcinoma; CP=chronic pancreatitis; NP=normal pancreas. (C) Serial sections of pancreatic cancer tissue stained for NFATc1 (upper panel) and calcineurin B (lower panel). Strong nuclear staining for NFATc1 in cancer cells was observed in 16 of 23 specimens. NFATc1-positive tumor cells consistently also stained positive for cytoplasmic and nuclear calcineurin B. Scale bars: 50 μm. (D) Expression levels of NFATc1 and calcineurin in pancreatic cancer cell lines as determined by RT–PCR (upper panel) and Western blot (lower panel) analyses. The human RPLP0 gene was used as housekeeping gene control for the human cell lines and the murine cyclophilin A gene (mPPIA) for the murine cell lines in the RT–PCR experiments. The Western blot for NFATc1 shows three major bands corresponding to different phosphorylation states of the protein. CnB=calcineurin B.
Figure 2
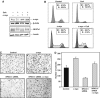
The Ca2+/calcineurin signaling pathway is active in pancreatic cancer cells. (A) Immunoblot analysis of subcellular localization of NFATc1 in pancreatic cancer cells. Panc-1 cells were either left untreated or treated with 1 μM CsA in the presence of serum for the indicated time intervals. Following subcellular fractionation, nuclear and cytoplasmic fractions were analyzed separately for the presence of NFATc1 protein by immunoblotting. (B) Immunocytological detection of NFATc1 localization in Panc-1 cells. Cells were kept in the absence of serum and either left untreated or treated with 1 μM CsA for 60 min, 1 μM ionomycin for 30 min, or a combination of CsA treatment followed by ionomycin treatment. Ionomycin stimulation led to a rapid and efficient translocation of NFATc1 (red signals) from the cytosol to the nucleus, which was completely blocked by CsA pretreatment. (C) DNA pulldown assays using oligonucleotides representing the consensus NFAT-binding sequence. Panc-1 cells were either left untreated or treated with 1 μM CsA for 30 min, 1 μM ionomycin for 15 min, or a combination of CsA treatment followed by ionomycin treatment. DNA–protein complexes from nuclear extracts were collected by precipitation with streptavidin–agarose beads and analyzed by Western blotting. Both, nuclear translocation (lower panel) as well as DNA-binding activity (upper panel) of NFATc1, were significantly enhanced by ionomycin, but completely blocked by CsA treatment. DNAP=DNA pulldown. (D) Activities of an NFAT-responsive reporter gene construct in Panc-1 cells. Cells were cotransfected with constitutively active (c.a.) calcineurin or dominant negative (d.n.) NFATc1 expression constructs and/or treated with 1 μM CsA or 1 μM ionomycin as indicated. Reporter gene activities are expressed as ‘fold activation' relative to untreated controls.
Figure 3
Inhibition of Ca2+/calcineurin signaling attenuates cell cycle progression and c_-myc_ expression in a subset of pancreatic cancer cell lines. (A) Proliferation assays demonstrating reduced growth of Panc-1 cells in response to calcineurin inhibitors. Cells were left untreated or treated with 1 μM CsA or 0.1 μM FK506 for 24 h as indicated. Proliferation was measured by [3H]thymidine incorporation assay. Data are representative of triplicate experiments and are displayed as bars+s.d. (B) Growth inhibition of Panc-1 cells by CsA is time- and dose-dependent. Panc-1 cells were grown for 24 or 48 h in the presence of different amounts of CsA as indicated. Proliferation was measured by [3H]thymidine incorporation assay. (C) Cell cycle analysis of pancreatic cancer cell lines. Cells were left untreated or treated with 1 μM CsA for 24 h and analyzed by propidium iodide staining and flow cytometry. The percentages of cells in the G1 and S phases, respectively, are indicated. CsA treatment resulted in cell cycle arrest, as indicated by a shift from the S to the G1 phase, in Panc-1 and ASPC-1 cells, but not in TD-2 or IMIM-PC2 cells. (D) Western blot analysis of c-myc protein expression in the pancreatic cancer cell lines. Cells were left untreated or treated with 1 μM CsA for 24 h as indicated. Total cell lysates were then analyzed for c-myc protein content using an anti-c-myc antibody. CsA treatment reduced c_-myc_ expression in Panc-1 and ASPC-1 cells, but not in TD-2 or IMIM-PC2 cells.
Figure 4
Anchorage-dependent and -independent cell growth and c_-myc_ activation in Ca2+/calcineurin-responsive cell lines is NFATc1-dependent. (A) Western blot analysis of NFATc1 knock down in Panc-1 cells. Cells were transiently transfected with three different NFATc1-specific siRNA sequences or a nonsilencing control siRNA and total cell lysates analyzed 48 h post-transfection using an anti-NFATc1 antibody. (B) Proliferation assays demonstrating dramatically reduced growth of Panc-1 cells after siRNA-mediated knock down of NFATc1. Proliferation was measured by [3H]thymidine incorporation assay. Data are representative of triplicate experiments and are displayed as bars+s.d. (C) Cell cycle analysis of NFATc1 knock-down cells. Cells were transiently transfected with NFATc1-specific siRNA sequences or nonsilencing control siRNA and analyzed 24 h post-transfection by propidium iodide staining and flow cytometry. The percentages of cells in the G1 and S phases, respectively, are indicated. NFATc1 knock down resulted in cell cycle arrest, as indicated by the shift from the S to the G1 phases. (D) Soft agar assays demonstrating significantly impaired anchorage-independent growth of NFATc1 knock-down cells. NFATc1 knock-down and control cells were seeded in soft agar 24 h post-transfection and the number of viable colonies was determined after 10 days. Data are representative of triplicate experiments and are displayed as bars+s.d. (E) Western blot analysis of the effect of NFATc1 knock down on c_-myc_ expression. Panc-1 cells were transiently transfected with the NFATc1-specific siRNA#2 or nonsilencing control siRNA and treated with 1 μM ionomycin for the indicated time periods. NFATc1 and c-myc protein was detected in total cell lysates using specific antibodies. Dephosphorylation, and thus activation, of NFATc1 by ionomycin is evidenced by the shift towards lower molecular weight bands (lower panel).
Figure 5
NFATc1-mediated growth promotion requires c_-myc_ activation. (A) Western blot analysis demonstrating c_-myc_ re-expression in CsA-treated Panc-1 cells. Cells were transiently transfected with a tetracycline-inducible c-myc expression construct or an empty vector control. Basal activity of the minimal promoter in the absence of tetracycline or doxycycline was sufficient to reconstitute basal c-myc expression levels in CsA-treated cells (lane 3). Analysis of nuclear extracts revealed that the inhibition of nuclear translocation of NFATc1 by CsA was not influenced by c-myc overexpression (lower panel). (B) Cell cycle analysis of cells re-expressing c-myc. CsA treatment significantly attenuated cell cycle progression in the control-transfected cells (upper panels), but had very little effect on the c-myc-transfected cells (lower panels). (C) Examples of soft agar assays demonstrating strong differences in colony formation after 10 days of incubation. Cells were left untransfected or transfected with the NFATc1-specific siRNA #2, the c-myc expression construct, or both, and the cells were seeded in soft agar 24 h post-transfection. Original magnification: × 100. (D) Statistical evaluation of soft agar assays. Reconstitution of c-myc expression completely abrogated the inhibitory effect of NFATc1 knock down on anchorage-independent growth of Panc-1 cells. Results are representative of triplicate experiments and are displayed as bars+s.d.
Figure 6
NFATc1 directly regulates c_-myc_ promoter activity. (A) Northern blot analysis demonstrating increased c_-myc_ transcription following transient transfection of Panc-1 cells with an NFATc1 expression construct. Loading control: 28S rRNA. (B) Activities of a reporter gene construct containing the full-length (2778 bp) c_-myc_ promoter sequence. Panc-1 cells were cotransfected with the reporter gene construct and increasing amounts of an NFATc1 expression construct or the NFATc1-specific siRNA#2 and treated with 1 μM CsA where indicated. Firefly luciferase reporter gene activities were normalized to Renilla luciferase activity and expressed as RLA. (C) Location of putative NFAT-binding sites (green bars) within the full-length c_-myc_ promoter fragment. The magnified section shows the sequence of the TGFβ inhibitory element (TIE) with the NFA-T binding site as well as previously described binding sites for the Smad-3, Sp1 and E2F transcription factors. (D) DNA pulldown assays using double-stranded oligonucleotides representing the wild-type (TIE-wt; upper sequence) or mutated (TIE-M1; mutation indicated in lower sequence) TIE. Panc-1 cells were grown in the absence of serum and either left untreated or treated with 1 μM ionomycin for 1 h as indicated. DNA–protein complexes were collected from nuclear extracts by precipitation with streptavidin–agarose beads and analyzed by immunoblotting using an anti-NFATc1 antibody. (E) Activities of reporter gene constructs containing TIE fragments with wild-type (TIE-wt) or mutated (TIE-M1) NFAT-binding sites. Panc-1 cells were cotransfected with the reporter gene constructs and NFATc1 expression vectors as indicated. Firefly luciferase reporter gene activities were normalized to Renilla luciferase activity and expressed as RLA. (F) Activities of reporter gene constructs containing the full-length (2778 bp) c_-myc_ promoter sequence carrying wild-type (c-myc-wt) or mutated (c-myc-M1) NFAT-binding sites within the TIE. Panc-1 cells were cotransfected with the reporter gene constructs and NFATc1 expression vectors as indicated. Firefly luciferase reporter gene activities were normalized to Renilla luciferase activity and expressed as RLA (for colour figure see online).
Similar articles
- Primers on molecular pathways--the NFAT transcription pathway in pancreatic cancer.
König A, Fernandez-Zapico ME, Ellenrieder V. König A, et al. Pancreatology. 2010;10(4):416-22. doi: 10.1159/000315035. Epub 2010 Aug 19. Pancreatology. 2010. PMID: 20720442 Free PMC article. Review. - Activation of the beta myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway.
Meissner JD, Umeda PK, Chang KC, Gros G, Scheibe RJ. Meissner JD, et al. J Cell Physiol. 2007 Apr;211(1):138-48. doi: 10.1002/jcp.20916. J Cell Physiol. 2007. PMID: 17111365 - NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells.
Köenig A, Linhart T, Schlengemann K, Reutlinger K, Wegele J, Adler G, Singh G, Hofmann L, Kunsch S, Büch T, Schäfer E, Gress TM, Fernandez-Zapico ME, Ellenrieder V. Köenig A, et al. Gastroenterology. 2010 Mar;138(3):1189-99.e1-2. doi: 10.1053/j.gastro.2009.10.045. Epub 2009 Nov 6. Gastroenterology. 2010. PMID: 19900447 Free PMC article. - Calcineurin/NFATc1 pathway contributes to cell proliferation in hepatocellular carcinoma.
Wang S, Kang X, Cao S, Cheng H, Wang D, Geng J. Wang S, et al. Dig Dis Sci. 2012 Dec;57(12):3184-8. doi: 10.1007/s10620-012-2255-8. Epub 2012 Jun 22. Dig Dis Sci. 2012. PMID: 22722879 - [Role of calcineurin/NFATc1 signaling pathway in cardiovascular development].
Wang X, Yan JC, Gong J. Wang X, et al. Zhonghua Xin Xue Guan Bing Za Zhi. 2012 Sep;40(9):798-800. Zhonghua Xin Xue Guan Bing Za Zhi. 2012. PMID: 23141098 Review. Chinese. No abstract available.
Cited by
- NFAT1 transcription factor regulates cell cycle progression and cyclin E expression in B lymphocytes.
Teixeira LK, Carrossini N, Sécca C, Kroll JE, DaCunha DC, Faget DV, Carvalho LD, de Souza SJ, Viola JP. Teixeira LK, et al. Cell Cycle. 2016 Sep;15(17):2346-59. doi: 10.1080/15384101.2016.1203485. Epub 2016 Jul 11. Cell Cycle. 2016. PMID: 27399331 Free PMC article. - The calcineurin/NFAT pathway is activated in diagnostic breast cancer cases and is essential to survival and metastasis of mammary cancer cells.
Quang CT, Leboucher S, Passaro D, Fuhrmann L, Nourieh M, Vincent-Salomon A, Ghysdael J. Quang CT, et al. Cell Death Dis. 2015 Feb 26;6(2):e1658. doi: 10.1038/cddis.2015.14. Cell Death Dis. 2015. PMID: 25719243 Free PMC article. - PPP3CB overexpression mediates EGFR TKI resistance in lung tumors via calcineurin/MEK/ERK signaling.
Gazzeri S, Zubchuk N, Montaudon E, Nemati F, Huot-Marchand S, Berardi G, Pucciarelli A, Dib Y, Nerini D, Oddou C, Pezet M, David-Boudet L, Ardin C, de Fraipont F, Maraver A, Girard N, Decaudin D, Toffart AC, Eymin B. Gazzeri S, et al. Life Sci Alliance. 2024 Oct 1;7(12):e202402873. doi: 10.26508/lsa.202402873. Print 2024 Dec. Life Sci Alliance. 2024. PMID: 39353739 Free PMC article. - Primers on molecular pathways--the NFAT transcription pathway in pancreatic cancer.
König A, Fernandez-Zapico ME, Ellenrieder V. König A, et al. Pancreatology. 2010;10(4):416-22. doi: 10.1159/000315035. Epub 2010 Aug 19. Pancreatology. 2010. PMID: 20720442 Free PMC article. Review. - Salt-inducible kinase 3 protects tumor cells from cytotoxic T-cell attack by promoting TNF-induced NF-κB activation.
Sorrentino A, Menevse AN, Michels T, Volpin V, Durst FC, Sax J, Xydia M, Hussein A, Stamova S, Spoerl S, Heuschneider N, Muehlbauer J, Jeltsch KM, Rathinasamy A, Werner-Klein M, Breinig M, Mikietyn D, Kohler C, Poschke I, Purr S, Reidell O, Martins Freire C, Offringa R, Gebhard C, Spang R, Rehli M, Boutros M, Schmidl C, Khandelwal N, Beckhove P. Sorrentino A, et al. J Immunother Cancer. 2022 May;10(5):e004258. doi: 10.1136/jitc-2021-004258. J Immunother Cancer. 2022. PMID: 35606086 Free PMC article.
References
- Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 - PubMed
- Al Daraji WI, Grant KR, Ryan K, Saxton A, Reynolds NJ (2002) Localization of calcineurin/NFAT in human skin and psoriasis and inhibition of calcineurin/NFAT activation in human keratinocytes by cyclosporin A. J Invest Dermatol 118: 779–788 - PubMed
- Amati B, Alevizopoulos K, Vlach J (1998) Myc and the cell cycle. Front Biosci 3: d250–d268 - PubMed
- Benedito AB, Lehtinen M, Massol R, Lopes UG, Kirchhausen T, Rao A, Bonni A (2005) The transcription factor NFAT3 mediates neuronal survival. J Biol Chem 280: 2818–2825 - PubMed
- Buchholz M, BieblA, Neesse A, Wagner M, Iwamura T, Leder G, Adler G, Gress TM (2003) SERPINE2 (protease nexin I) promotes extracellular matrix production and local invasion of pancreatic tumors in vivo. Cancer Res 63: 4945–4951 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous