Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling - PubMed (original) (raw)
Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling
Céline DerMardirossian et al. Mol Biol Cell. 2006 Nov.
Abstract
Rho GTPases (Rac, Rho, and Cdc42) play important roles in regulating cell function through their ability to coordinate the actin cytoskeleton, modulate the formation of signaling reactive oxidant species, and control gene transcription. Activation of Rho GTPase signaling pathways requires the regulated release of Rho GTPases from RhoGDI complexes, followed by their reuptake after membrane cycling. We show here that Src kinase binds and phosphorylates RhoGDI both in vitro and in vivo at Tyr156. Analysis of Rho GTPase-RhoGDI complexes using in vitro assays of complexation and in vivo by coimmunoprecipitation analysis indicates that Src-mediated phosphorylation of Tyr156 causes a dramatic decrease in the ability of RhoGDI to form a complex with RhoA, Rac1, or Cdc42. Phosphomimetic mutation of Tyr156-->Glu results in the constitutive association of RhoGDI(Y156E) with the plasma membrane and/or associated cortical actin. Substantial cortical localization of tyrosine-phosphorylated RhoGDI is also observed in fibroblasts expressing active Src, where it is most evident in podosomes and regions of membrane ruffling. Expression of membrane-localized RhoGDI(Y156E) mutant is associated with enhanced cell spreading and membrane ruffling. These results suggest that Src-mediated RhoGDI phosphorylation is a novel physiological mechanism for regulating Rho GTPase cytosol membrane-cycling and activity.
Figures
Figure 1.
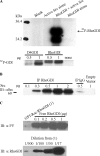
Src phosphorylates uncomplexed RhoGDI at Tyrosine 156 in vitro. (A) Src phosphorylates RhoGDI directly. Recombinant RhoGDI (2 μg) purified from E. coli was subjected to in vitro kinase assay using recombinant active Src (1 μg), as in Materials and Methods (top panels). Phosphorylation of the indicated amounts of D4GDI versus RhoGDI were compared under the same conditions (bottom panels). (B) Interaction of His-tagged RhoGDIwt with overexpressed active Src. HeLa cells were cotransfected with His-tagged RhoGDI wt and different amounts of active Src Y527F cDNA, as indicated. RhoGDI was then immunoprecipitated and probed with Src antibody for coimmunoprecipitated Src proteins. The IgG IP control represents a nonspecific rabbit IgG pulldown in the presence of 0.5 μg Src plasmid DNA. (C) Src does not phosphorylate Rac-RhoGDI complex in vitro. (A) GST-Rac1–RhoGDI complex immobilized on glutathione beads and recombinant purified uncomplexed RhoGDI were subjected to an in vitro kinase assay using active recombinant Src, and then tyrosine phosphorylation was determined by blotting with mAb 4G10, as in Materials and Methods. The top panel shows that only free RhoGDI, but not RhoGDI complexed with Rac, is tyrosine phosphorylated by Src. The bottom panel shows a RhoGDI immunoblot of various dilutions (indicated in parentheses) of the GST-Rac1–RhoGDI complex used in the kinase assay of the top panel (undiluted = 1).
Figure 2.
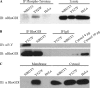
Src phosphorylates RhoGDI at Tyr156 in vivo. HeLa cells were cotransfected with active Src Y527F (or empty vector) and the indicated mutant versions of His-tagged RhoGDI. The top panel shows equal amounts of each construct were immunoprecipitated. Immunoblot of immunoprecipitated RhoGDI constructs with 4G10 phosphotyrosine antibody indicate that RhoGDI is primarily phosphorylated by Src at Tyrosine 156.
Figure 3.
Endogenous RhoGDI is tyrosine-phosphorylated in Src3T3 cells and is found associated with the membrane fraction. (A) Whole cell lysates prepared from SrcY527F-expressing NIH-3T3 cells (Y527F) or control NIH-3T3 cells versus Hela cell controls were immunoblotted for RhoGDI (αRhoGDI) either in the whole lysate (right panels) or after immunoprecipitation with 4G10 phospho-tyrosine antibody, as in Materials and Methods. (B) Lysates from SrcY527F-expressing NIH-3T3 cells or control NIH-3T3 cells were immunoprecipitated with RhoGDI antibody (left lanes) or unspecific rabbit IgG (middle lanes) and then immunoblotted for RhoGDI and phospho-tyrosine. Four and 10 μg of total cytosol protein were loaded as controls (right lanes). (C) Membrane versus cytosol fractions were prepared from SrcY527F-expressing NIH-3T3 cells or control NIH-3T3 cells versus Hela cell controls as described in Materials and Methods, and then the fractions were immunoblotted for RhoGDI. Results shown here are representative of at least two experiments.
Figure 4.
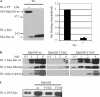
Tyrosine-phosphorylated RhoGDI and a RhoGDI phosphomimetic mutant exhibit less affinity for Rac, Rho, and Cdc42. (A) Myc-tagged Rac1 was pulled down from cell lysates by recombinant GST-RhoGDI immobilized on glutathione beads either without (−) or with (+) in vitro phosphorylation by recombinant active Src (left panel), as in Materials and Methods. Pulldowns were immunoblotted for both phospho-tyrosine with 4G10 (αPY) and the presence of associated Rac1 with myc mAb 9E10 (αMyc). The results from three experiments were quantified (right panel) and are shown as mean ± SE. (B) Myc-tagged Rac1-, RhoA-, and Cdc42-expresing cell lysates were incubated with the indicated amounts (in micrograms) of recombinant GST-RhoGDI constructs immobilized on glutathione beads and then pulled down by centrifugation, washed, and analyzed. Substitution in RhoGDI of tyrosine 156 with glutamic acid (Y156E), but not with phenylalanine (Y156F), mimics the results observed with Src-phosphorylated RhoGDI in A by reducing complex formation with all three Rho GTPases at all levels of added GDI protein. (C) The ability of the indicated pure recombinant RhoGDI constructs to extract GST-Rac1 from Sf9 cell membranes was determined, as in Materials and Methods. There was a small amount of Rac1 that became soluble even in the absence of RhoGDI (shown as − control). Results shown above are typical of at least three independent experiments.
Figure 5.
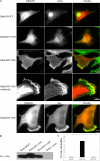
Tyrosine–phosphorylated RhoGDI enhances and localizes to membrane ruffles. (A) HeLa cells were transfected with either His-tagged RhoGDI wt (WT), His-tagged RhoGDI Y156F (Y156F), or His-tagged RhoGDI Y156E (Y156E). At 6 h after transfection, the cells were fixed and stained for His tag (red) and F-actin (green). Arrowheads indicate ruffles prominently containing His-RhoGDI Y156E. Colocalization of RhoGDI Y156E with F-actin in membrane ruffles was verified by confocal microscopy (lower panels, as indicated), as in Materials and Methods. Colocalization of His-RhoGDI Y156E (red) with Rac1 (green) in membrane ruffles was also evident (lowermost panels) and was also confirmed by confocal microscopy (unpublished data). Bar, 10 μm or 5 μm (confocal), as indicated. Results shown are typical of more than three independent experiments. (B) RhoGDI immunoblots showing approximately equivalent levels of transfected RhoGDI expression in the cells used in A. (C) The percentage of cells expressing various RhoGDI mutants that exhibited a membrane ruffling phenotype was quantified. Results shown are from n = 272 cells in three separate experiments and are given as mean ± SE.
Figure 6.
RhoGDI localizes to Src-dependent rosettes in Src3T3 fibroblasts. SrcY527F-expressing NIH-3T3 cells were analyzed by immunofluorescence, as described in Materials and Methods. (A) Cells were either untreated (−SU6656) or treated with 10 μM SU6656 (+SU6656) overnight before analysis for localization of F-actin (blue; antibody at 1:10 dilution), phospho-tyrosine (green; antibody at 1:250 dilution), and RhoGDI (red; antibody at 1:250 dilution), as indicated. The rightmost panel is a threefold magnification of the boxed area indicated in the overlay. (B) Localization of F-actin (blue; antibody at 1:10 dilution), vinculin (green; antibody at 1:1000 dilution), and RhoGDI (red; antibody at 1:250 dilution), as indicated. (C) Localization of F-actin (blue; antibody at 1:10 dilution), RhoA (green; antibody at 1:100 dilution), and RhoGDI (red; antibody at 1:250 dilution), as indicated. In each case, the areas of colocalization for all three proteins are shown as white (Pearson's r ≥ 0.7). The lowermost panels are a threefold magnification of the boxed area indicated in the lower magnification actin panel.
Similar articles
- Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase.
DerMardirossian C, Schnelzer A, Bokoch GM. DerMardirossian C, et al. Mol Cell. 2004 Jul 2;15(1):117-27. doi: 10.1016/j.molcel.2004.05.019. Mol Cell. 2004. PMID: 15225553 - An activating mutant of Rac1 that fails to interact with Rho GDP-dissociation inhibitor stimulates membrane ruffling in mammalian cells.
Gandhi PN, Gibson RM, Tong X, Miyoshi J, Takai Y, Konieczkowski M, Sedor JR, Wilson-Delfosse AL. Gandhi PN, et al. Biochem J. 2004 Mar 1;378(Pt 2):409-19. doi: 10.1042/BJ20030979. Biochem J. 2004. PMID: 14629200 Free PMC article. - RhoGDI: multiple functions in the regulation of Rho family GTPase activities.
Dovas A, Couchman JR. Dovas A, et al. Biochem J. 2005 Aug 15;390(Pt 1):1-9. doi: 10.1042/BJ20050104. Biochem J. 2005. PMID: 16083425 Free PMC article. Review. - Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling.
Olofsson B. Olofsson B. Cell Signal. 1999 Aug;11(8):545-54. doi: 10.1016/s0898-6568(98)00063-1. Cell Signal. 1999. PMID: 10433515 Review.
Cited by
- Semaphorin 5A and plexin-B3 inhibit human glioma cell motility through RhoGDIalpha-mediated inactivation of Rac1 GTPase.
Li X, Lee AY. Li X, et al. J Biol Chem. 2010 Oct 15;285(42):32436-45. doi: 10.1074/jbc.M110.120451. Epub 2010 Aug 9. J Biol Chem. 2010. PMID: 20696765 Free PMC article. - RhoGDI3 and RhoG: Vesicular trafficking and interactions with the Sec3 Exocyst subunit.
Morin A, Cordelières FP, Cherfils J, Olofsson B. Morin A, et al. Small GTPases. 2010 Nov;1(3):142-156. doi: 10.4161/sgtp.1.3.15112. Small GTPases. 2010. PMID: 21686268 Free PMC article. - RhoGDIbeta-induced hypertrophic growth in H9c2 cells is negatively regulated by ZAK.
Huang CY, Yang LC, Liu KY, Liao PH, Chou JI, Chou MY, Lin WW, Yang JJ. Huang CY, et al. J Biomed Sci. 2009 Jan 22;16(1):11. doi: 10.1186/1423-0127-16-11. J Biomed Sci. 2009. PMID: 19272173 Free PMC article. - α2β1 integrins spatially restrict Cdc42 activity to stabilise adherens junctions.
Howden JD, Michael M, Hight-Warburton W, Parsons M. Howden JD, et al. BMC Biol. 2021 Jun 23;19(1):130. doi: 10.1186/s12915-021-01054-9. BMC Biol. 2021. PMID: 34158053 Free PMC article.
References
- Abram C. L., Seals D. F., Pass I., Salinsky D., Maurer L., Roth T. M., Courtneidge S. A. The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J. Biol. Chem. 2003;278:16844–16851. - PubMed
- Benzing T. Signaling at the slit diaphragm. J. Am. Soc. Nephrol. 2004;15:1382–1391. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous