Caveolin-1-deficient mice show defects in innate immunity and inflammatory immune response during Salmonella enterica serovar Typhimurium infection - PubMed (original) (raw)
Caveolin-1-deficient mice show defects in innate immunity and inflammatory immune response during Salmonella enterica serovar Typhimurium infection
Freddy A Medina et al. Infect Immun. 2006 Dec.
Abstract
A number of studies have shown an association of pathogens with caveolae. To this date, however, there are no studies showing a role for caveolin-1 in modulating immune responses against pathogens. Interestingly, expression of caveolin-1 has been shown to occur in a regulated manner in immune cells in response to lipopolysaccharide (LPS). Here, we sought to determine the role of caveolin-1 (Cav-1) expression in Salmonella pathogenesis. Cav-1(-/-) mice displayed a significant decrease in survival when challenged with Salmonella enterica serovar Typhimurium. Spleen and tissue burdens were significantly higher in Cav-1(-/-) mice. However, infection of Cav-1(-/-) macrophages with serovar Typhimurium did not result in differences in bacterial invasion. In addition, Cav-1(-/-) mice displayed increased production of inflammatory cytokines, chemokines, and nitric oxide. Regardless of this, Cav-1(-/-) mice were unable to control the systemic infection of Salmonella. The increased chemokine production in Cav-1(-/-) mice resulted in greater infiltration of neutrophils into granulomas but did not alter the number of granulomas present. This was accompanied by increased necrosis in the liver. However, Cav-1(-/-) macrophages displayed increased inflammatory responses and increased nitric oxide production in vitro in response to Salmonella LPS. These results show that caveolin-1 plays a key role in regulating anti-inflammatory responses in macrophages. Taken together, these data suggest that the increased production of toxic mediators from macrophages lacking caveolin-1 is likely to be responsible for the marked susceptibility of caveolin-1-deficient mice to S. enterica serovar Typhimurium.
Figures
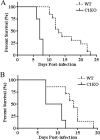
FIG. 1.
Cav-1−/− mice display significantly reduced survival rates upon challenge with a highly virulent serovar Typhimurium strain. Cav-1+/+ (WT, wild type) and Cav-1−/− (C1KO, Cav-1 knockout) mice were administered 1 × 103 CFU i.v (A) or 104 CFU of p.o. (B) and monitored daily for survival (n ≥ 9). Survival curves were analyzed with the log rank test and revealed statistically significant differences for mice challenged both i.v and p.o. (P ≤ 0.001).
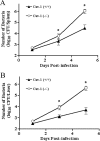
FIG. 2.
Numbers of live serovar Typhimurium organisms in the spleens or livers of Cav-1+/+ and Cav-1−/− mice after bacterial challenge. Cav-1+/+ (n = 5) and Cav-1−/− mice (n = 5) were infected i.v. with 1 × 103 CFU of serovar Typhimurium, and bacterial counts from spleen (A) and liver (B) homogenates were determined by culture on agar plates at days 1, 3, and 5 postinfection. Data are the mean number of CFU ± standard error of the mean. Differences were considered statistically significant at P values of ≤0.05.
FIG. 3.
Dramatic increase of serum cytokine production in Cav-1−/− mice in response to serovar Typhimurium infection. Mice were bled after 3 days of challenge with serovar Typhimurium and the serum chemokine (A) and cytokine (B) levels of the proinflammatory cytokines and the anti-inflammatory cytokines were measured. Data are the mean ± standard error of the mean (n ≥ 7). Differences were considered statistically significant at P values of ≤0.05. WT, Cav-1+/+ (wild type); C1KO, Cav-1−/− (knockout).
FIG. 4.
Increased production of serum nitric oxide in Cav-1−/− mice in response to serovar Typhimurium infection. Mice were bled after 3 days of challenge with serovar Typhimurium, and the serum nitric oxide levels were determined (n ≥ 7). Differences were considered statistically significant at P values of ≤0.05.
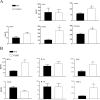
FIG. 5.
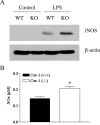
Increased production of inflammatory cytokines from LPS-stimulated Cav-1−/− macrophages. Macrophages from Cav-1+/+ (WT, wild type) and Cav-1−/− (KO, knockout) mice were isolated by peritoneal lavage. Cav-1+/+ and Cav-1−/− macrophages were cultured with 1 μg/ml of serovar Typhimurium for 24 h. (A) Western blot analysis of caveolin-1 expression levels are shown. β-actin was employed as an equal loading control. Note the increase in caveolin-1 expression in response to LPS in Cav-1+/+ macrophages. (B and C) Concentrations of chemokines and cytokines in the culture supernatants were measured by LINCOplex. Data are the mean ± standard error of the mean from triplicate wells. Differences were considered statistically significant at P values of ≤0.05.
FIG. 6.
Increased nitric oxide production and iNOS activity in Cav-1−/− macrophages. Macrophages from Cav-1+/+ (WT, wild type) and Cav-1−/− (KO, knockout) mice were isolated by peritoneal lavage. Cav-1+/+ and Cav-1−/− macrophages were cultured with 1 μg/ml of serovar Typhimurium for 24 h, and nitric oxide levels were measured or iNOS expression was assessed from lysates. Data are the mean ± standard error of the mean from triplicate wells. Differences were considered statistically significant at P values of ≤0.05.
FIG. 7.
Reduced STAT3 phosphorylation in Cav-1−/− macrophages. Cav-1+/+ (WT, wild type) and Cav-1−/− (KO, knockout) macrophages were cultured with 1 μg/ml of serovar Typhimurium for 0, 0.5, 1, 3, and 24 h. Lysates from stimulated macrophages were tested for pSTAT3, stripped, and reprobed for STAT3.
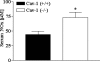
FIG. 8.
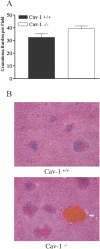
Granuloma burden and histopathological evaluation of liver section stained by H-E. (A) Liver granulomas from Cav-1+/+ and Cav-1−/− mice (n ≥ 5) infected with 1 × 103 CFU serovar Typhimurium were counted in 25 light microscopic fields. Data are the mean ± standard error of the mean. Statistical analysis by a Student's t test revealed that there was no statistically significant difference. (B) Cav-1−/− mice show an increased amount of liver necrosis at 3 days postinfection.
FIG. 9.
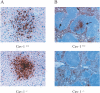
Neutrophil recruitment in serovar Typhimurium-infected Cav-1+/+ and Cav-1−/− mice. (A) Cav-1−/− mice show an increase in neutrophil recruitment in liver granulomas at 3 days postinfection. (B) Spleen sections from serovar Typhimurium-infected Cav-1+/+ mice show neutrophil infiltration in the white pulp, while sections from Cav-1−/− mice lack any such infiltration.
Similar articles
- Toll-Like receptor 2 (TLR2) and TLR9 play opposing roles in host innate immunity against Salmonella enterica serovar Typhimurium infection.
Zhan R, Han Q, Zhang C, Tian Z, Zhang J. Zhan R, et al. Infect Immun. 2015 Apr;83(4):1641-9. doi: 10.1128/IAI.02870-14. Epub 2015 Feb 9. Infect Immun. 2015. PMID: 25667264 Free PMC article. - Murein lipoprotein is a critical outer membrane component involved in Salmonella enterica serovar typhimurium systemic infection.
Fadl AA, Sha J, Klimpel GR, Olano JP, Niesel DW, Chopra AK. Fadl AA, et al. Infect Immun. 2005 Feb;73(2):1081-96. doi: 10.1128/IAI.73.2.1081-1096.2005. Infect Immun. 2005. PMID: 15664952 Free PMC article. - Proteins from latex of Calotropis procera prevent septic shock due to lethal infection by Salmonella enterica serovar Typhimurium.
Lima-Filho JV, Patriota JM, Silva AF, Filho NT, Oliveira RS, Alencar NM, Ramos MV. Lima-Filho JV, et al. J Ethnopharmacol. 2010 Jun 16;129(3):327-34. doi: 10.1016/j.jep.2010.03.038. Epub 2010 Apr 3. J Ethnopharmacol. 2010. PMID: 20371281 - Critical role of type I interferon-induced macrophage necroptosis during infection with Salmonella enterica serovar Typhimurium.
Liang S, Qin X. Liang S, et al. Cell Mol Immunol. 2013 Mar;10(2):99-100. doi: 10.1038/cmi.2012.68. Epub 2012 Dec 24. Cell Mol Immunol. 2013. PMID: 23262973 Free PMC article. Review. No abstract available. - Salmonella and the Inflammasome: Battle for Intracellular Dominance.
Crowley SM, Knodler LA, Vallance BA. Crowley SM, et al. Curr Top Microbiol Immunol. 2016;397:43-67. doi: 10.1007/978-3-319-41171-2_3. Curr Top Microbiol Immunol. 2016. PMID: 27460804 Review.
Cited by
- Caveolin-1 protects B6129 mice against Helicobacter pylori gastritis.
Hitkova I, Yuan G, Anderl F, Gerhard M, Kirchner T, Reu S, Röcken C, Schäfer C, Schmid RM, Vogelmann R, Ebert MP, Burgermeister E. Hitkova I, et al. PLoS Pathog. 2013;9(4):e1003251. doi: 10.1371/journal.ppat.1003251. Epub 2013 Apr 11. PLoS Pathog. 2013. PMID: 23592983 Free PMC article. - Caveolin-1 modifies the immunity to Pseudomonas aeruginosa.
Gadjeva M, Paradis-Bleau C, Priebe GP, Fichorova R, Pier GB. Gadjeva M, et al. J Immunol. 2010 Jan 1;184(1):296-302. doi: 10.4049/jimmunol.0900604. Epub 2009 Nov 30. J Immunol. 2010. PMID: 19949109 Free PMC article. - Effects of Probiotic Enterococcus faecium from Yak on the Intestinal Microflora and Metabolomics of Mice with Salmonella Infection.
Ran X, Li X, Xie X, Lei J, Yang F, Chen D. Ran X, et al. Probiotics Antimicrob Proteins. 2024 Jun;16(3):1036-1051. doi: 10.1007/s12602-023-10102-5. Epub 2023 Jun 5. Probiotics Antimicrob Proteins. 2024. PMID: 37273089 - Role of Caveolin Proteins in Sepsis.
Sowa G. Sowa G. Pediatr Ther. 2012;2012(Suppl 2):001. doi: 10.4172/2161-0665.S2-001. Epub 2012 Jan 12. Pediatr Ther. 2012. PMID: 26618071 Free PMC article. - Caveolae as Potential Hijackable Gates in Cell Communication.
Dudãu M, Codrici E, Tanase C, Gherghiceanu M, Enciu AM, Hinescu ME. Dudãu M, et al. Front Cell Dev Biol. 2020 Oct 27;8:581732. doi: 10.3389/fcell.2020.581732. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33195223 Free PMC article. Review.
References
- Catron, D. M., M. D. Sylvester, Y. Lange, M. Kadekoppala, B. D. Jones, D. M. Monack, S. Falkow, and K. Haldar. 2002. The Salmonella-containing vacuole is a major site of intracellular cholesterol accumulation and recruits the GPI-anchored protein CD55. Cell Microbiol. 4:315-328. - PubMed
- Centers for Disease Control and Prevention. 1999. Summary of notifiable diseases, United States, 1998. Morb. Mortal. Wkly. Rep. 47:ii-92. - PubMed
- Chan, S. Y., C. J. Empig, F. J. Welte, R. F. Speck, A. Schmaljohn, J. F. Kreisberg, and M. A. Goldsmith. 2001. Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell 106:117-126. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases