Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish - PubMed (original) (raw)
Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish
Laurence Covassin et al. Dev Biol. 2006.
Abstract
In this study, we utilize fluorescent activated cell sorting (FACS) of cells from transgenic zebrafish coupled with microarray analysis to globally analyze expression of cell type specific genes. We find that it is possible to isolate cell populations from Tg(fli1:egfp)(y1) zebrafish embryos that are enriched in vascular, hematopoietic and pharyngeal arch cell types. Microarray analysis of GFP+ versus GFP- cells isolated from Tg(fli1:egfp)(y1) embryos identifies genes expressed in hematopoietic, vascular and pharyngeal arch tissue, consistent with the expression of the fli1:egfp transgene in these cell types. Comparison of expression profiles from GFP+ cells isolated from embryos at two different time points reveals that genes expressed in different fli1+ cell types display distinct temporal expression profiles. We also demonstrate the utility of this approach for gene discovery by identifying numerous previously uncharacterized genes that we find are expressed in fli1:egfp-positive cells, including new markers of blood, endothelial and pharyngeal arch cell types. In parallel, we have developed a database to allow easy access to both our microarray and in situ results. Our results demonstrate that this is a robust approach for identification of cell type specific genes as well as for global analysis of cell type specific gene expression in zebrafish embryos.
Figures
Figure 1
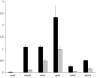
Enrichment of GFP+ cells from Tg(fli1:egfp) y1 zebrafish embryos. A-D. Diagnostic fluorescence activated cell sorting of dissociated Tg(fli1:egfp) y1 embryos. A. Cells from Tg(fli1:egfp) y1 embryos. B. Cells from non-transgenic wild type embryos. A, B. P3 and P4 demarcate cells sorted as GFP− and GFP+, respectively. C. GFP− cells following FACS isolation. D. GFP+ cells following FACS isolation. E. Relative expression of indicated genes in GFP+ (black bars) and GFP− (grey bars) determined by quantitative RT-PCR. Relative expression levels were determined by normalization to_ef1a_.
Figure 2
Quantitative RT-PCR analysis of previously described non-fli1 + and ubiquitously-expressed genes (see Table 2). qRT-PCR was performed on RNA derived from fli1:egfp + (black bars) and _egfp_− (grey bars) cells from 22–24 hpf embryos.
Figure 3
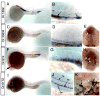
Whole mount in situ hybridization analysis of blood cell markers. A, B.Dr.1382 at 26 hpf; box indicate magnified view in B. B. Arrows indicate expression in erythroid cells; nc – notochord. C-E. Dr.30834 at 26 hpf; box indicates magnified view in D. E. Dorsal view of head, anterior is up; white arrow indicates expression in the heart. F-H. Dr.919 at 24 hpf. F. Box indicates magnified view in G. H. Dorsal view of the head, anterior is up. White arrows denote expression in white blood cells on yolk sac. I-K.Dr.6172 at 24 hpf. I. Black box indicates magnified view in J, white box indicates view in K. J. expression in floor plate (fp), hypochord (hc), and a blood cell (bc). K. Blood cells expressing Dr.6172 indicated with black arrows. White arrows denote adjacent cells with similar morphology that do not express Dr.6172. A-D, F, G, I, J. Lateral view, dorsal is up anterior to the left.
Figure 4
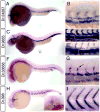
Whole mount in situ hybridization analysis of cardiovascular markers. A, B.Dr.24287 at 24 hpf; box indicates magnified view in B. B. Black bracket indicates dorsal aorta, white bracket shows posterior cardinal vein. C-E. Dr. 9655. C. Embryo at 30 hpf. D. Magnified view of dorsal aorta (red bracket) and posterior cardinal vein (white bracket) at 24 hpf; E. Dorsal aorta (red bracket) and posterior cardinal vein (white bracket) at 30 hpf. F, G. Dr.33656. G. expression in segmental arteries (black arrow) and dorsal aorta (red bracket); expression is absent from posterior cardinal vein (white bracket). H, I. Dr.398 at 24 hpf. H. Arrow indicated expression in heart; inset head-on view showing heart expression (arrow). I. Magnified view of embryo in H.
Figure 5
Whole mount in situ hybridization analysis of pharyngeal arch markers. A, B. Dr.51180 at 26 hpf; arrow indicates expression in trunk blood vessels B. Dorsal view of head of embryo in A; arrows indicate expression in pharyngeal arches. C, D. BI671621. D. Dorsal view of head of embryo in C; arrows indicate expression in pharyngeal arches.
Figure 6
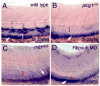
plcg1 and mib are required for artery specific expression of Dr.33656. A. Wild type embryo. B. Embryo mutant for plcg1 y18. C. Embryo mutant for mib ta52b. D. Embryo injected with 5 ng Rbpsuh morpholino. Lateral views, anterior to the left, dorsal is up. A-D. White arrow indicates expression in pronephric ducts (out of focal plane). Red bracket demarcates dorsal aorta in A.-D. and white bracket indicates posterior cardinal vein in A.
Figure 7
Notch represses dab2 and vsg1 expression. A-E. dab2 expression. A. Wild type sibling embryo following heat shock. Red bracket indicates dorsal aorta, white bracket posterior cardinal vein. B. Tg(hsp70:gal4) kca4;Tg(uas:myc-notch-intra) kca3 embryo following heat shock. Myc-positive cells in the neural tube are indicated by black arrowheads. C, D. Dorsal view of_dab2_ expression in pronephric ducts (white arrows). C. Same embryo as shown in A; D. same embryos as seen in B. E. Ectopic_dab2_ expression in the dorsal aorta (white bracket) of a_mib_ ta52b mutant embryo. F-H. vsg1 expression. F. Wild type sibling embryo following heat shock. Red bracket indicates dorsal aorta, white bracket posterior cardinal vein. G. Tg(hsp70:gal4) kca4;Tg(uas:myc-notch-intra) kca3 embryo following heat shock. Myc-positive cells in the neural tube are indicated by black arrowheads. H. Ectopic vsg1 expression in the dorsal aorta (white bracket) of a mib ta52b mutant embryo.
Figure 8
A Zebrafish microarray database. A. Opening page of database with possible pairwise combinations and user defined analysis. B. Results output page showing Accession number, gene name, Unigene, and values from selected microarray experiment. Also includes links to in situ data. C. Example of and individual gene page with values across all available microarray data.
Similar articles
- Autotaxin-Lysophosphatidic Acid Axis Acts Downstream of Apoprotein B Lipoproteins in Endothelial Cells.
Gibbs-Bar L, Tempelhof H, Ben-Hamo R, Ely Y, Brandis A, Hofi R, Almog G, Braun T, Feldmesser E, Efroni S, Yaniv K. Gibbs-Bar L, et al. Arterioscler Thromb Vasc Biol. 2016 Oct;36(10):2058-67. doi: 10.1161/ATVBAHA.116.308119. Epub 2016 Aug 25. Arterioscler Thromb Vasc Biol. 2016. PMID: 27562917 - Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants.
Sumanas S, Jorniak T, Lin S. Sumanas S, et al. Blood. 2005 Jul 15;106(2):534-41. doi: 10.1182/blood-2004-12-4653. Epub 2005 Mar 31. Blood. 2005. PMID: 15802528 Free PMC article. - Fli1+ cells transcriptional analysis reveals an Lmo2-Prdm16 axis in angiogenesis.
Matrone G, Xia B, Chen K, Denvir MA, Baker AH, Cooke JP. Matrone G, et al. Proc Natl Acad Sci U S A. 2021 Aug 3;118(31):e2008559118. doi: 10.1073/pnas.2008559118. Proc Natl Acad Sci U S A. 2021. PMID: 34330825 Free PMC article. - Transgenic fluorescent zebrafish Tg(fli1:EGFP)y¹ for the identification of vasotoxicity within the zFET.
Delov V, Muth-Köhne E, Schäfers C, Fenske M. Delov V, et al. Aquat Toxicol. 2014 May;150:189-200. doi: 10.1016/j.aquatox.2014.03.010. Epub 2014 Mar 19. Aquat Toxicol. 2014. PMID: 24685623 - Global gene expression analysis of single cells.
Kamme F, Erlander MG. Kamme F, et al. Curr Opin Drug Discov Devel. 2003 Mar;6(2):231-6. Curr Opin Drug Discov Devel. 2003. PMID: 12669459 Review.
Cited by
- Competition between Jagged-Notch and Endothelin1 Signaling Selectively Restricts Cartilage Formation in the Zebrafish Upper Face.
Barske L, Askary A, Zuniga E, Balczerski B, Bump P, Nichols JT, Crump JG. Barske L, et al. PLoS Genet. 2016 Apr 8;12(4):e1005967. doi: 10.1371/journal.pgen.1005967. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27058748 Free PMC article. - G protein-coupled receptor 183 facilitates endothelial-to-hematopoietic transition via Notch1 inhibition.
Zhang P, He Q, Chen D, Liu W, Wang L, Zhang C, Ma D, Li W, Liu B, Liu F. Zhang P, et al. Cell Res. 2015 Oct;25(10):1093-107. doi: 10.1038/cr.2015.109. Epub 2015 Sep 11. Cell Res. 2015. PMID: 26358189 Free PMC article. - Dual Roles of Fer Kinase Are Required for Proper Hematopoiesis and Vascular Endothelium Organization during Zebrafish Development.
Dunn EM, Billquist EJ, VanderStoep AL, Bax PG, Westrate LM, McLellan LK, Peterson SC, MacKeigan JP, Putzke AP. Dunn EM, et al. Biology (Basel). 2017 Nov 23;6(4):40. doi: 10.3390/biology6040040. Biology (Basel). 2017. PMID: 29168762 Free PMC article. - Antioxidant, Pro-Survival and Pro-Regenerative Effects of Conditioned Medium from Wharton's Jelly Mesenchymal Stem Cells on Developing Zebrafish Embryos.
Reina C, Cardella C, Lo Pinto M, Pucci G, Acuto S, Maggio A, Cavalieri V. Reina C, et al. Int J Mol Sci. 2023 Aug 25;24(17):13191. doi: 10.3390/ijms241713191. Int J Mol Sci. 2023. PMID: 37685998 Free PMC article.
References
- Amsterdam A. Insertional mutagenesis in zebrafish. Dev Dyn. 2003;228:523–34. - PubMed
- Barrallo A, Gonzalez-Sarmiento R, Garcia-Isidoro M, et al. Differential brain expression of a new beta-actin gene from zebrafish (Danio rerio) Eur J Neurosci. 1999;11:369–72. - PubMed
- Claudio JO, Zhu YX, Benn SJ, et al. HACS1 encodes a novel SH3-SAM adaptor protein differentially expressed in normal and malignant hematopoietic cells. Oncogene. 2001;20:5373–7. - PubMed
- Crosier PS, Bardsley A, Horsfield JA, et al. In situ hybridization screen in zebrafish for the selection of genes encoding secreted proteins. Dev Dyn. 2001;222:637–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK032520/DK/NIDDK NIH HHS/United States
- R03 HD047490/HD/NICHD NIH HHS/United States
- 5 P30 DK32520/DK/NIDDK NIH HHS/United States
- 5R03HD047490/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous