Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences - PubMed (original) (raw)
Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences
Derrick E Fouts. Nucleic Acids Res. 2006.
Abstract
Phage_Finder, a heuristic computer program, was created to identify prophage regions in completed bacterial genomes. Using a test dataset of 42 bacterial genomes whose prophages have been manually identified, Phage_Finder found 91% of the regions, resulting in 7% false positive and 9% false negative prophages. A search of 302 complete bacterial genomes predicted 403 putative prophage regions, accounting for 2.7% of the total bacterial DNA. Analysis of the 285 putative attachment sites revealed tRNAs are targets for integration slightly more frequently (33%) than intergenic (31%) or intragenic (28%) regions, while tmRNAs were targeted in 8% of the regions. The most popular tRNA targets were Arg, Leu, Ser and Thr. Mapping of the insertion point on a consensus tRNA molecule revealed novel insertion points on the 5' side of the D loop, the 3' side of the anticodon loop and the anticodon. A novel method of constructing phylogenetic trees of phages and prophages was developed based on the mean of the BLAST score ratio (BSR) of the phage/prophage proteomes. This method verified many known bacteriophage groups, making this a useful tool for predicting the relationships of prophages from bacterial genomes.
Figures
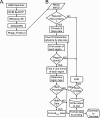
Figure 1
Flow chart of Phage_Finder pipeline (A) and Phage_Finder.pl script (B) logic. Standard symbols for constructing flow charts were used.
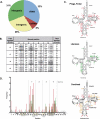
Figure 2
Predicted prophage target-site distributions. The distribution of targets where Phage_Finder found putative attachment sites (A). The genetic code table indicates the distribution of tRNA targets (B). For each codon, the number of phages from Williams, 2002 and the number of predicted prophages from this study are indicated, separated by a colon. The gray-highlighted numbers demarcate those codons that are targeted six or more times. The point of insertion on a consensus tRNA molecule was mapped (C) for Phage_Finder predicted prophages (upper), phages and prophages from the literature [(27), middle] and the two datasets combined (lower). The arrows point to the nucleotide insertion point while the numbers indicate number of insertions at each insertion point. Red arrows and numbers in the combined dataset show those locations that are unique to either dataset, while gold colored arrows and numbers highlight common insertion points between the two datasets. The frequency and position of insertion into a consensus tRNA gene is noted in (D). Red bars indicate _Phage_Finder_-predicted insertion events while green bars represent insertion events reported from the literature (27).
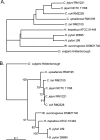
Figure 3
Test phylogenetic tree generated by converting the BSR of BLASTP bidirectional matches into distance (A). Whole genome BLASTP data from Fouts et al., 2005 (37) was used to compute this tree. The previously published 16S rRNA tree (B) is shown for comparison.
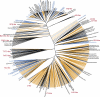
Figure 4
Phylogenetic analysis of Phage_Finder predicted prophages, known prophages and sequenced phage genomes. The radial tree was constructed with branch length extensions. The branches were colored as follows: sequenced phage genomes (black), known prophages (blue), Phage_Finder predicted prophage regions (gold). Only key phages or prophages are noted for clarity. Known phage groups are indicated in red.
Similar articles
- Prophage insertion sites.
Campbell A. Campbell A. Res Microbiol. 2003 May;154(4):277-82. doi: 10.1016/S0923-2508(03)00071-8. Res Microbiol. 2003. PMID: 12798232 - The impact of prophages on bacterial chromosomes.
Canchaya C, Fournous G, Brüssow H. Canchaya C, et al. Mol Microbiol. 2004 Jul;53(1):9-18. doi: 10.1111/j.1365-2958.2004.04113.x. Mol Microbiol. 2004. PMID: 15225299 Review. - Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences.
Bose M, Barber RD. Bose M, et al. In Silico Biol. 2006;6(3):223-7. In Silico Biol. 2006. PMID: 16922685 - Genomic organization and molecular analysis of the inducible prophage EJ-1, a mosaic myovirus from an atypical pneumococcus.
Romero P, López R, García E. Romero P, et al. Virology. 2004 May 1;322(2):239-52. doi: 10.1016/j.virol.2004.01.029. Virology. 2004. PMID: 15110522 - Comparative genomics of phages and prophages in lactic acid bacteria.
Desiere F, Lucchini S, Canchaya C, Ventura M, Brüssow H. Desiere F, et al. Antonie Van Leeuwenhoek. 2002 Aug;82(1-4):73-91. Antonie Van Leeuwenhoek. 2002. PMID: 12369206 Review.
Cited by
- Uncovering the prevalence and diversity of integrating conjugative elements in actinobacteria.
Ghinet MG, Bordeleau E, Beaudin J, Brzezinski R, Roy S, Burrus V. Ghinet MG, et al. PLoS One. 2011;6(11):e27846. doi: 10.1371/journal.pone.0027846. Epub 2011 Nov 16. PLoS One. 2011. PMID: 22114709 Free PMC article. - PHISDetector: A Tool to Detect Diverse In Silico Phage-host Interaction Signals for Virome Studies.
Zhou F, Gan R, Zhang F, Ren C, Yu L, Si Y, Huang Z. Zhou F, et al. Genomics Proteomics Bioinformatics. 2022 Jun;20(3):508-523. doi: 10.1016/j.gpb.2022.02.003. Epub 2022 Mar 8. Genomics Proteomics Bioinformatics. 2022. PMID: 35272051 Free PMC article. - DEPhT: a novel approach for efficient prophage discovery and precise extraction.
Gauthier CH, Abad L, Venbakkam AK, Malnak J, Russell DA, Hatfull GF. Gauthier CH, et al. Nucleic Acids Res. 2022 Jul 22;50(13):e75. doi: 10.1093/nar/gkac273. Nucleic Acids Res. 2022. PMID: 35451479 Free PMC article. - Tight regulation of the intS gene of the KplE1 prophage: a new paradigm for integrase gene regulation.
Panis G, Duverger Y, Courvoisier-Dezord E, Champ S, Talla E, Ansaldi M. Panis G, et al. PLoS Genet. 2010 Oct 7;6(10):e1001149. doi: 10.1371/journal.pgen.1001149. PLoS Genet. 2010. PMID: 20949106 Free PMC article. - Isolation, identification, and whole-genome sequencing of high-yield protease bacteria from Daqu of ZhangGong Laojiu.
Liu Y, Fu J, Wang L, Zhao Z, Wang H, Han S, Sun X, Pan C. Liu Y, et al. PLoS One. 2022 Apr 26;17(4):e0264677. doi: 10.1371/journal.pone.0264677. eCollection 2022. PLoS One. 2022. PMID: 35472204 Free PMC article.
References
- Sullivan M.B., Waterbury J.B., Chisholm S.W. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047–1051. - PubMed
- Mann N.H., Cook A., Millard A., Bailey S., Clokie M. Bacterial photosynthesis genes in a virus. Nature. 2003;424:741. - PubMed
- Stone R. Bacteriophage therapy. Stalin's forgotten cure. Science. 2002;298:728–731. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials