SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily - PubMed (original) (raw)
SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily
Ricardo DeMarco et al. BMC Evol Biol. 2006.
Abstract
Background: The CACTA (also called En/Spm) superfamily of DNA-only transposons contain the core sequence CACTA in their Terminal Inverted Repeats (TIRs) and so far have only been described in plants. Large transcriptome and genome sequence data have recently become publicly available for Schistosoma mansoni, a digenetic blood fluke that is a major causative agent of schistosomiasis in humans, and have provided a comprehensive repository for the discovery of novel genes and repetitive elements. Despite the extensive description of retroelements in S. mansoni, just a single DNA-only transposon belonging to the Merlin family has so far been reported in this organism.
Results: We describe a novel S. mansoni transposon named SmTRC1, for S. mansoni Transposon Related to CACTA 1, an element that shares several characteristics with plant CACTA transposons. Southern blotting indicates approximately 30-300 copies of SmTRC1 in the S. mansoni genome. Using genomic PCR followed by cloning and sequencing, we amplified and characterized a full-length and a truncated copy of this element. RT-PCR using S. mansoni mRNA followed by cloning and sequencing revealed several alternatively spliced transcripts of this transposon, resulting in distinct ORFs coding for different proteins. Interestingly, a survey of complete genomes from animals and fungi revealed several other novel TRC elements, indicating new families of DNA transposons belonging to the CACTA superfamily that have not previously been reported in these kingdoms. The first three bases in the S. mansoni TIR are CCC and they are identical to those in the TIRs of the insects Aedes aegypti and Tribolium castaneum, suggesting that animal TRCs may display a CCC core sequence.
Conclusion: The DNA-only transposable element SmTRC1 from S. mansoni exhibits various characteristics, such as generation of multiple alternatively-spliced transcripts, the presence of terminal inverted repeats at the extremities of the elements flanked by direct repeats and the presence of a Transposase_21 domain, that suggest a distant relationship to CACTA transposons from Magnoliophyta. Several sequences from other Metazoa and Fungi code for proteins similar to those encoded by SmTRC1, suggesting that such elements have a common ancestry, and indicating inheritance through vertical transmission before separation of the Eumetazoa, Fungi and Plants.
Figures
Figure 1
SmTRC1 elements. A: Agarose gel electrophoresis of typical PCR amplification products of S. mansoni genomic DNA with primers designed from the sequence of SmTRC1 extremities. B: Schematic representation of the SmTRC1 element derived in silico from S. mansoni shotgun genomic sequencing and assembly data obtained from the Sanger Institute (Supercontig 0018735) or from direct sequencing of clones amplified by PCR from genomic DNA obtained in this work (SmTRC1f1 and SmTRC1d1). Black boxes indicate the Terminal Inverted Repeats (TIR). Light gray boxes indicate the predicted SmTRC1-ORF and the dark gray box indicates the Transposase_21 domain within this ORF. The hatched box indicates a region with tandem repeats.
Figure 2
Transposon inverted and direct repeats. A: the complete sequence of SmTRC1 TIR is shown in this panel. Dots represent the transposon sequence not shown in the figure. B: blue boxes show direct repeats flanking S. mansoni and other animal TIRs (in gray). Only part of the S. mansoni TIR is represented in the figure. The dots represent the transposon sequence not shown in the figure. C: examples of target-site duplication created upon SmTRC1 insertion. Examples of alignments of sequences flanking SmTRC1 insertions (S-0000026, S-0000464 and S-0000144) with paralogous genomic sequences lacking transposon insertions (BH202398.1, BN000802.1 and AL620357.1) that were found in the S. mansoni public sequences database. The paralogous "gap" sequence (marked as –) presumably corresponds to the genomic target sequence before a transposon insertion event. Blue boxes indicate the target-site duplication in the flanking sequence. The number on the side of each sequence represents the supercontig from which it was derived (in the case of transposon inserted sequences) or GenBank accession numbers (in the case of paralogous sequences). D: TIR sequences from diverse CACTA superfamily animal and plant elements. The regions with high and medium levels of identity among the sequences are shown as black and gray columns, respectively.
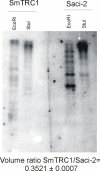
Figure 3
Southern blotting of SmTRC1. S. mansoni genomic DNA (5 μg) digested with the indicated restriction enzyme was loaded in each lane and analyzed by Southern blotting with a specific radiolabeled probe for SmTRC1. A parallel experiment was run with a probe for the Saci-2 retrotransposon [18], which was used as a benchmark. Probes of similar sizes and the same number of radioactive counts were used for each of the two hybridizations. Below the figure, the ratio between the total intensities of the SmTRC1/Saci-2 signals is indicated. This value was calculated for each digestion by integrating the signal from all the bands, and the average and total deviation was obtained by computing data from the two different digestions.
Figure 4
Alternatively spliced forms of SmTRC1 transcripts. A: agarose gel electrophoresis of products from an RT-PCR reaction with S. mansoni mRNA using primers designed from the sequence of the extremities of previously deposited ESTs mapping to the full-length SmTRC1 sequence. The "no RT" lane indicates a control in which no reverse transcriptase was added to the reaction medium. B: full-length SmTRC1 sequence (top scheme) and relative mapping positions of five existing ESTs from GenBank (Accession numbers shown next to each) and of three newly sequenced transcripts obtained by cloning the major band derived from the RT-PCR shown in panel A (Clones B1, B2 and B4 as indicated). Black boxes in the top scheme indicate the Terminal Inverted Repeats (TIR); light gray boxes indicate a predicted SmTRC1-ORF and the dark gray box indicates a Transposase_21 domain within this ORF; the hatched box indicates a region with tandem repeats. Thin black bars below the top scheme indicate mapped exons derived from each transcript; a white box indicates a region of a particular transcript not mapped to this specific copy of the SmTRC1 genomic sequence. Thin continuous lines represent junctions between interconnected exons in the transcripts, defining an intron with the canonical GT-AG splicing sites. Dashed lines represent junctions between interconnected exons in the transcripts, defining an intron without the canonical GT-AG splicing sites. Two "A"s indicate the presence of a poly-A tail. C: schematic representation of 3 clones of SmTRC1 transcripts. The scale in this part of the figure is expanded in comparison to that used in part B above. Light gray boxes indicate predicted ORF. Names inside the boxes indicate different hypothetical protein products coded by those transcripts. The asterisk indicates a stop codon present in the transcript but not in the equivalent genomic sequence of the full-length SmTRC1 element.
Figure 5
Multiple alignment of the Transposase_21 domains of proteins of CACTA related transposons from diverse organisms. Typical plant CACTA transposon sequences from six Magnoliophyta were included in the alignment. In addition, eleven novel CACTA-related elements identified here were included: seven from Eumetazoa and four from Fungi. Shading indicates the level of conservation of each residue. Boxes with Roman numbers I to III indicate conserved motifs of the Transposase_21 domain in all organisms. Box marked with A indicates a Transposase_21 motif displayed only by Eumetazoa and Fungi proteins.
Figure 6
Phylogenetic tree for the Transposase_21 domains of CACTA-like transposases. The tree was constructed by the neighbor-joining method using the three conserved regions indicated by boxes I to III in Figure 5 and excluding positions with gaps. Numbers represent the confidence of the branches assigned by bootstrap analysis (in 1,000 samplings); bootstrap values lower than 500 are omitted from the figure. The names indicate a transposon member of the CACTA or of the
T
ransposon
R
elated to
C
ACTA (TRC) family belonging to the organism indicated. Circles indicate the 3 different proposed families of transposons within the CACTA superfamily.
Similar articles
- Curupira-1 and Curupira-2, two novel Mutator-like DNA transposons from the genomes of human parasites Schistosoma mansoni and Schistosoma japonicum.
Jacinto DS, Muniz Hdos S, Venancio TM, Wilson RA, Verjovski-Almeida S, Demarco R. Jacinto DS, et al. Parasitology. 2011 Aug;138(9):1124-33. doi: 10.1017/S0031182011000886. Epub 2011 Jul 15. Parasitology. 2011. PMID: 21756422 - Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences.
Feschotte C. Feschotte C. Mol Biol Evol. 2004 Sep;21(9):1769-80. doi: 10.1093/molbev/msh188. Epub 2004 Jun 9. Mol Biol Evol. 2004. PMID: 15190130 - Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum.
Chalvet F, Grimaldi C, Kaper F, Langin T, Daboussi MJ. Chalvet F, et al. Mol Biol Evol. 2003 Aug;20(8):1362-75. doi: 10.1093/molbev/msg155. Epub 2003 May 30. Mol Biol Evol. 2003. PMID: 12777515 - Transposons in filamentous fungi--facts and perspectives.
Kempken F, Kück U. Kempken F, et al. Bioessays. 1998 Aug;20(8):652-9. doi: 10.1002/(SICI)1521-1878(199808)20:8<652::AID-BIES8>3.0.CO;2-K. Bioessays. 1998. PMID: 9841641 Review. - Assembly of the Tc1 and mariner transposition initiation complexes depends on the origins of their transposase DNA binding domains.
Brillet B, Bigot Y, Augé-Gouillou C. Brillet B, et al. Genetica. 2007 Jun;130(2):105-20. doi: 10.1007/s10709-006-0025-2. Epub 2006 Aug 16. Genetica. 2007. PMID: 16912840 Review.
Cited by
- Remobilization of Tol2 transposons in Xenopus tropicalis.
Yergeau DA, Kelley CM, Kuliyev E, Zhu H, Sater AK, Wells DE, Mead PE. Yergeau DA, et al. BMC Dev Biol. 2010 Jan 22;10:11. doi: 10.1186/1471-213X-10-11. BMC Dev Biol. 2010. PMID: 20096115 Free PMC article. - Abundant degenerate miniature inverted-repeat transposable elements in genomes of epichloid fungal endophytes of grasses.
Fleetwood DJ, Khan AK, Johnson RD, Young CA, Mittal S, Wrenn RE, Hesse U, Foster SJ, Schardl CL, Scott B. Fleetwood DJ, et al. Genome Biol Evol. 2011;3:1253-64. doi: 10.1093/gbe/evr098. Epub 2011 Sep 26. Genome Biol Evol. 2011. PMID: 21948396 Free PMC article. - Idaten is a new cold-inducible transposon of Volvox carteri that can be used for tagging developmentally important genes.
Ueki N, Nishii I. Ueki N, et al. Genetics. 2008 Nov;180(3):1343-53. doi: 10.1534/genetics.108.094672. Epub 2008 Sep 14. Genetics. 2008. PMID: 18791222 Free PMC article. - Repeated horizontal transfers of four DNA transposons in invertebrates and bats.
Tang Z, Zhang HH, Huang K, Zhang XG, Han MJ, Zhang Z. Tang Z, et al. Mob DNA. 2015 Jan 17;6(1):3. doi: 10.1186/s13100-014-0033-1. eCollection 2015. Mob DNA. 2015. PMID: 25606061 Free PMC article. - A genome survey of Moniliophthora perniciosa gives new insights into Witches' Broom Disease of cacao.
Mondego JM, Carazzolle MF, Costa GG, Formighieri EF, Parizzi LP, Rincones J, Cotomacci C, Carraro DM, Cunha AF, Carrer H, Vidal RO, Estrela RC, García O, Thomazella DP, de Oliveira BV, Pires AB, Rio MC, Araújo MR, de Moraes MH, Castro LA, Gramacho KP, Gonçalves MS, Neto JP, Neto AG, Barbosa LV, Guiltinan MJ, Bailey BA, Meinhardt LW, Cascardo JC, Pereira GA. Mondego JM, et al. BMC Genomics. 2008 Nov 18;9:548. doi: 10.1186/1471-2164-9-548. BMC Genomics. 2008. PMID: 19019209 Free PMC article.
References
- Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, D.C. , ASM Press; 2002. p. xviii, 1204 p., [32] p. of plates.
- Capy P, Bazin C, Higuet D, Langin T. Molecular biology intelligence unit. Austin, Tex; New York , Landes Bioscience ; North American distributor Chapman & Hall; 1998. Dynamics and evolution of transposable elements; p. 197 p..
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials