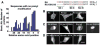PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane - PubMed (original) (raw)
PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane
Won Do Heo et al. Science. 2006.
Abstract
Many signaling, cytoskeletal, and transport proteins have to be localized to the plasma membrane (PM) in order to carry out their function. We surveyed PM-targeting mechanisms by imaging the subcellular localization of 125 fluorescent protein-conjugated Ras, Rab, Arf, and Rho proteins. Out of 48 proteins that were PM-localized, 37 contained clusters of positively charged amino acids. To test whether these polybasic clusters bind negatively charged phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] lipids, we developed a chemical phosphatase activation method to deplete PM PI(4,5)P2. Unexpectedly, proteins with polybasic clusters dissociated from the PM only when both PI(4,5)P2 and phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] were depleted, arguing that both lipid second messengers jointly regulate PM targeting.
Figures
Fig. 1
A survey of the subcellular localization of 125 small GTPases shows that most PM-localized small GTPases have targeting motifs with clusters of polybasic amino acids. (A) Confocal images of the subcellular localization of CFP-conjugated small GTPases in NIH3T3 cells (full set of images in NIH3T3 and HeLa cells in fig. S1). The four main PM-targeting motifs are represented in the image panels. (B) Correlation between PM localization and the presence of lysine residues in a region 5 to 20 amino acids from the C terminus. (C) Phylogenetic tree of 48 small GTPases that were identified to be partially or fully localized to the PM. Individual membrane targeting elements are color coded: red for polybasic clusters and blue, green, and orange for palmitoyl, prenyl, and myristoyl consensus sequences, respectively. (D) Twenty-amino-acid-long C-terminal tail fragments of Rit and KRas are PM-localized. Lack of PM targeting of a KRas tail fragment without the polybasic region (right). Scale bars, 10 μm.
Fig. 2
Depletion of PI(4,5)P2 and PI(3,4,5)P3 dissociates Rit, Rin, and MARCKS ED with polybasic-nonlipid targeting motifs from the PM. (A) Development of a chemically inducible translocation method to deplete PI(4,5)P2 from the inner leaflet of the PM (22, 23). The PI(4,5)P2 biosensor YFP-PLCd-PH was cotransfected to monitor depletion of PI(4,5)P2. (B) Depletion of PI(4,5)P2 caused only a small reduction in the PM localization of YFP-conjugated Rit tail. (C) A small, but significant, PDGF receptor–mediated increase in Rit tail PM localization can be reversed by addition of the PI3-kinase inhibitor LY29 (before and 9 min after addition of 5 μM iRap). The bar graph shows a quantification of the same experiment. The PM dissociation index is the relative ratio of internal over PM fluorescence; that is, **F1cyt/F**1PM * **F0PM/F**0cyt, with **F**0 and **F**1 as the fluorescent intensities before and after PI(4,5)P2 and PI(3,4,5)P3 depletion. (D) Joint reduction in PI(4,5)P2 and PI(3,4,5)P3 triggered a near-complete dissociation of Rit tail and MARCKS ED from the PM. (E) Quantitative analysis of the CF-Inp and/or LY29-triggered PM dissociation of MARCKS ED, and Rit and Rin tails. PLCδ-PH and HRas tails are shown as controls. The inactive LY29 analog LY30 was used as a control (24). Scale bars, 10 μm.
Fig. 3
Polybasic subclusters and hydrophobic amino acids are required for PM targeting by polybasic-nonlipid targeting motifs. (A) Statistical analysis of the relative sequence location of positively charged amino acids in nonprenylated small GTPases. (B) Loss of PM targeting of Rit tail fragment after removal of a single subcluster with positively charged amino acids. (C) Identification of a tryptophan residue in the polybasic regions of Rit and Rin that mediates PM over nuclear localization. Full-length small GTPases, as well as tail fragment mutants, are shown. (D) Identification of two additional hydrophobic amino acid residues that contribute to the PM targeting of Rit. Scale bars, 10 μm.
Fig. 4
Depletion of PI(4,5)P2 and PI(3,4,5)P3 dissociates proteins with polybasic-myristoyl and polybasic-prenyl targeting motifs from the PM. (A) Aligned sequences of the N terminus (6 amino acids) and C terminus (20 amino acids) of six ARF family members. Confocal images of an Arl7 mutant construct lacking the N-terminal myristoylation motif (left), a C-terminal tagged Arl7 control construct (middle), and a mutant construct with an internal CFP tag and a flexible C-terminal polybasic Arl7 tail (right). (B) PM localization motifs in geranylgeranylated Rab35. Confocal images from left to right: localization of wild-type Rab35, wild-type tail fragment, wild-type tail lacking geranylgeranylation motif (ΔCC), Rab35 tail ΔCC mutant with KRas caax (Rab35 tail caax), and Rab35 tail caax mutant lacking polybasic amino acids (Rab35 tail caax ΔpB). (C) Depletion of PI(4,5)P2 by CF-Inp activation without L29 addition causes only a minimal PM dissociation of Arl7, KRas tail fragment, and Rab35. (D) Quantitative analysis of the much larger PM dissociation of polybasic-lipid modified proteins after depletion of PI(4,5)P2 and PI(3,4,5)P3 by CF-Inp activation and addition of LY29. Scale bars, 10 μm.
Comment in
- Cell biology. Tools to tamper with phosphoinositides.
McLaughlin S. McLaughlin S. Science. 2006 Dec 1;314(5804):1402-3. doi: 10.1126/science.1136314. Epub 2006 Nov 9. Science. 2006. PMID: 17095656 No abstract available.
Similar articles
- Cell biology. Tools to tamper with phosphoinositides.
McLaughlin S. McLaughlin S. Science. 2006 Dec 1;314(5804):1402-3. doi: 10.1126/science.1136314. Epub 2006 Nov 9. Science. 2006. PMID: 17095656 No abstract available. - RAB-10-GTPase-mediated regulation of endosomal phosphatidylinositol-4,5-bisphosphate.
Shi A, Liu O, Koenig S, Banerjee R, Chen CC, Eimer S, Grant BD. Shi A, et al. Proc Natl Acad Sci U S A. 2012 Aug 28;109(35):E2306-15. doi: 10.1073/pnas.1205278109. Epub 2012 Aug 6. Proc Natl Acad Sci U S A. 2012. PMID: 22869721 Free PMC article. - Regulation of podosomes by integrin alphavbeta3 and Rho GTPase-facilitated phosphoinositide signaling.
Chellaiah MA. Chellaiah MA. Eur J Cell Biol. 2006 Apr;85(3-4):311-7. doi: 10.1016/j.ejcb.2006.01.008. Epub 2006 Feb 7. Eur J Cell Biol. 2006. PMID: 16460838 Review. - Interactions between Rab and Arf GTPases regulate endosomal phosphatidylinositol-4,5-bisphosphate during endocytic recycling.
Shi A, Grant BD. Shi A, et al. Small GTPases. 2013 Apr-Jun;4(2):106-9. doi: 10.4161/sgtp.23477. Epub 2013 Feb 7. Small GTPases. 2013. PMID: 23392104 Free PMC article. - PI3K class IB pathway in neutrophils.
Andrews S, Stephens LR, Hawkins PT. Andrews S, et al. Sci STKE. 2007 Oct 9;2007(407):cm3. doi: 10.1126/stke.4072007cm3. Sci STKE. 2007. PMID: 17925574 Review.
Cited by
- Endoplasmic Reticulum-Plasma Membrane Contact Sites as an Organizing Principle for Compartmentalized Calcium and cAMP Signaling.
Crul T, Maléth J. Crul T, et al. Int J Mol Sci. 2021 Apr 29;22(9):4703. doi: 10.3390/ijms22094703. Int J Mol Sci. 2021. PMID: 33946838 Free PMC article. Review. - Regulation of voltage-dependent calcium channels by RGK proteins.
Yang T, Colecraft HM. Yang T, et al. Biochim Biophys Acta. 2013 Jul;1828(7):1644-54. doi: 10.1016/j.bbamem.2012.10.005. Epub 2012 Oct 10. Biochim Biophys Acta. 2013. PMID: 23063948 Free PMC article. Review. - Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae.
Chen RE, Thorner J. Chen RE, et al. Biochim Biophys Acta. 2007 Aug;1773(8):1311-40. doi: 10.1016/j.bbamcr.2007.05.003. Epub 2007 May 22. Biochim Biophys Acta. 2007. PMID: 17604854 Free PMC article. Review. - A novel ROP/RAC GTPase effector integrates plant cell form and pattern formation.
Bloch D, Hazak O, Lavy M, Yalovsky S. Bloch D, et al. Plant Signal Behav. 2008 Jan;3(1):41-3. doi: 10.4161/psb.3.1.4838. Plant Signal Behav. 2008. PMID: 19704766 Free PMC article. - Directly light-regulated binding of RGS-LOV photoreceptors to anionic membrane phospholipids.
Glantz ST, Berlew EE, Jaber Z, Schuster BS, Gardner KH, Chow BY. Glantz ST, et al. Proc Natl Acad Sci U S A. 2018 Aug 14;115(33):E7720-E7727. doi: 10.1073/pnas.1802832115. Epub 2018 Jul 31. Proc Natl Acad Sci U S A. 2018. PMID: 30065115 Free PMC article.
References
- Fivaz M, Meyer T. Neuron. 2003;40:319. - PubMed
- Teruel MN, Meyer T. Cell. 2000;103:181. - PubMed
- Guan JL. Science. 2004;303:773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH064801/MH/NIMH NIH HHS/United States
- R01 GM063702/GM/NIGMS NIH HHS/United States
- R01 GM030179-24A1/GM/NIGMS NIH HHS/United States
- R01 GM030179/GM/NIGMS NIH HHS/United States
- R01 GM030179-25/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous