p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans - PubMed (original) (raw)
p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans
Emily R Troemel et al. PLoS Genet. 2006.
Abstract
The PMK-1 p38 mitogen-activated protein kinase pathway and the DAF-2-DAF-16 insulin signaling pathway control Caenorhabditis elegans intestinal innate immunity. pmk-1 loss-of-function mutants have enhanced sensitivity to pathogens, while daf-2 loss-of-function mutants have enhanced resistance to pathogens that requires upregulation of the DAF-16 transcription factor. We used genetic analysis to show that the pathogen resistance of daf-2 mutants also requires PMK-1. However, genome-wide microarray analysis indicated that there was essentially no overlap between genes positively regulated by PMK-1 and DAF-16, suggesting that they form parallel pathways to promote immunity. We found that PMK-1 controls expression of candidate secreted antimicrobials, including C-type lectins, ShK toxins, and CUB-like genes. Microarray analysis demonstrated that 25% of PMK-1 positively regulated genes are induced by Pseudomonas aeruginosa infection. Using quantitative PCR, we showed that PMK-1 regulates both basal and infection-induced expression of pathogen response genes, while DAF-16 does not. Finally, we used genetic analysis to show that PMK-1 contributes to the enhanced longevity of daf-2 mutants. We propose that the PMK-1 pathway is a specific, indispensable immunity pathway that mediates expression of secreted immune response genes, while the DAF-2-DAF-16 pathway appears to regulate immunity as part of a more general stress response. The contribution of the PMK-1 pathway to the enhanced lifespan of daf-2 mutants suggests that innate immunity is an important determinant of longevity.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
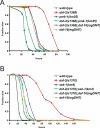
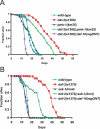
Figure 1. The PMK-1 p38 MAPK Pathway Is Required for daf-2 Enhanced Pathogen Resistance
(A) pmk-1(km25) suppresses daf-2(e1368) enhanced pathogen resistance. (B) sek-1(km4) suppresses daf-2(e1370) enhanced pathogen resistance. Slow killing assays were performed with P. aeruginosa strain PA14 under standard conditions, and each graph shows the average of three plates for each strain, with each plate containing 20–30 worms. Results are representative of 3 independent assays. See Table S1 for more assays and statistical analysis.
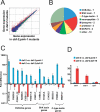
Figure 2. PMK-1 and DAF-16 Upregulate Distinct Genes
(A) Intensity plot of genes upregulated by PMK-1 in daf-2(e1368) mutant. Plot represents signal intensity of all 22,500 sequences on Affymetrix C. elegans GeneChip. The _x_-axis shows expression level in daf-2(e1368);pmk-1(km25) animals and the _y_-axis shows expression level in daf-2(e1368) animals. Sequences colored red are considered upregulated by wild-type PMK-1 and sequences colored green are considered downregulated by wild-type PMK-1 (using Resolver; p <0.01). Diagonal lines delineate a 2-fold change. See Table S2 for a complete list of genes differentially expressed between daf-2 and daf-2;pmk-1. (B) Pie chart of gene classes upregulated by PMK-1. The top 36 genes induced are represented. (C and D) qRT-PCR comparing expression in daf-2(e1368) versus daf-2(e1368);pmk-1(km25), and daf-2(e1368) versus daf-2(e1368);daf-16(mgDf47). Results are the average of two biological replicates, each replicate measured in duplicate and normalized to a control gene. Error bars are SEM. (C) shows genes upregulated by PMK-1 and (D) shows genes upregulated by DAF-16.
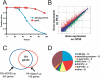
Figure 3. P. aeruginosa Induces Expression of Genes in Similar Gene Classes as PMK-1–Induced Genes
(A) PA14 slow killing assay comparing survival of fer-15;fem-1 young adult animals on gacA and wild-type PA14. Animals were raised in the same way as for harvest for microarrays. (B) Intensity plot of genes regulated by PA14 versus OP50 at 4 h. Sequences colored red are considered upregulated by PA14 and sequences colored green are considered downregulated by wild-type PA14 (using Resolver; p <0.01). Diagonal lines delineate a 2-fold change. See Table S4 for a complete list of genes differentially expressed between PA14 and OP50. (C) Venn diagram of overlap between genes induced in PA14 versus OP50 at 4 h comparison and PA14 versus gacA at 4 h comparison. (D) Pie chart of gene classes induced by PA14 versus OP50. The genes (59) that are upregulated greater than 5-fold are represented.
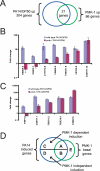
Figure 4. PMK-1 Regulates Basal and Inducible Expression of P. aeruginosa–Induced Genes
(A) Venn diagram of overlap between genes regulated by PMK-1 and P. aeruginosa. (B) (C) qRT-PCR analysis of PA14-induced gene expression in wild-type animals and in pmk-1 mutants. Results are the average of two biological replicates, each replicate measured in duplicate and normalized to a control gene. Error bars are SEM. (D) Diagram of different gene classes regulated by PMK-1 and/or P. aeruginosa. PMK-1 is required for basal and inducible regulations of class A genes. PMK-1 is required for basal, but not inducible expression of class B genes. PMK-1 is required for inducible but not basal expression of class C genes. PMK-1 is required for neither basal nor inducible expression of class D genes. PMK-1 regulates basal expression of class E genes, but these genes are not induced by P. aeruginosa.
Figure 5. The PMK-1 p38 MAPK Pathway Is Partially Required for Increased Longevity of daf-2
(A) pmk-1(km25) partially suppresses enhanced longevity of daf-2(e1368). (B) sek-1(km4) partially suppresses enhanced longevity of daf-2(e1370). See Table S7 for quantitative data.
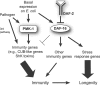
Figure 6. A Model for Regulation of Immunity/Longevity by the PMK-1 p38 MAPK Pathway and the DAF-2–DAF-16 Insulin-Signaling Pathway in C. elegans
We propose that the PMK-1 p38 MAPK pathway plays an active role in response to bacterial pathogens. PMK-1 controls basal levels of pathogen response genes on E. coli and also mediates induction of these genes upon infection. In contrast, DAF-16 controls a separate group of immune genes and does not mediate the infection response, but does turn down expression of some pathogen-induced genes. Certain pathogen response genes are induced independently of PMK-1 or DAF-16, indicating the existence of a third pathway. PMK-1 is required for the enhanced pathogen resistance of daf-2 mutants because it is the predominant pathway contributing to immunity. Finally, we suggest that PMK-1 contributes to enhanced longevity of daf-2 mutants because intact immune function promotes longevity.
Similar articles
- Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans.
Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, Whitney JK, Kamanzi O, Matsumoto K, Hisamoto N, Kim DH. Shivers RP, et al. PLoS Genet. 2010 Apr 1;6(4):e1000892. doi: 10.1371/journal.pgen.1000892. PLoS Genet. 2010. PMID: 20369020 Free PMC article. - The Caenorhabditis elegans germ line regulates distinct signaling pathways to control lifespan and innate immunity.
Alper S, McElwee MK, Apfeld J, Lackford B, Freedman JH, Schwartz DA. Alper S, et al. J Biol Chem. 2010 Jan 15;285(3):1822-8. doi: 10.1074/jbc.M109.057323. Epub 2009 Nov 18. J Biol Chem. 2010. PMID: 19923212 Free PMC article. - PMK-1 p38 MAPK promotes cadmium stress resistance, the expression of SKN-1/Nrf and DAF-16 target genes, and protein biosynthesis in Caenorhabditis elegans.
Keshet A, Mertenskötter A, Winter SA, Brinkmann V, Dölling R, Paul RJ. Keshet A, et al. Mol Genet Genomics. 2017 Dec;292(6):1341-1361. doi: 10.1007/s00438-017-1351-z. Epub 2017 Aug 1. Mol Genet Genomics. 2017. PMID: 28766017 Free PMC article. - Regulation of aging and innate immunity in C. elegans.
Kurz CL, Tan MW. Kurz CL, et al. Aging Cell. 2004 Aug;3(4):185-93. doi: 10.1111/j.1474-9728.2004.00108.x. Aging Cell. 2004. PMID: 15268752 Review. - Transcriptional responses to pathogens in Caenorhabditis elegans.
Shivers RP, Youngman MJ, Kim DH. Shivers RP, et al. Curr Opin Microbiol. 2008 Jun;11(3):251-6. doi: 10.1016/j.mib.2008.05.014. Epub 2008 Jun 21. Curr Opin Microbiol. 2008. PMID: 18567532 Free PMC article. Review.
Cited by
- C. elegans aversive olfactory learning generates diverse intergenerational effects.
Pereira AG, Gracida X, Kagias K, Zhang Y. Pereira AG, et al. J Neurogenet. 2020 Sep-Dec;34(3-4):378-388. doi: 10.1080/01677063.2020.1819265. Epub 2020 Sep 17. J Neurogenet. 2020. PMID: 32940103 Free PMC article. - Cytoplasmic LSM-1 protein regulates stress responses through the insulin/IGF-1 signaling pathway in Caenorhabditis elegans.
Cornes E, Porta-De-La-Riva M, Aristizábal-Corrales D, Brokate-Llanos AM, García-Rodríguez FJ, Ertl I, Díaz M, Fontrodona L, Reis K, Johnsen R, Baillie D, Muñoz MJ, Sarov M, Dupuy D, Cerón J. Cornes E, et al. RNA. 2015 Sep;21(9):1544-53. doi: 10.1261/rna.052324.115. Epub 2015 Jul 6. RNA. 2015. PMID: 26150554 Free PMC article. - Does Candida albicans Als5p amyloid play a role in commensalism in Caenorhabditis elegans?
Bois M, Singh S, Samlalsingh A, Lipke PN, Garcia MC. Bois M, et al. Eukaryot Cell. 2013 May;12(5):703-11. doi: 10.1128/EC.00020-13. Epub 2013 Mar 8. Eukaryot Cell. 2013. PMID: 23475704 Free PMC article. - Genetics of aging in Caenorhabditis elegans.
Antebi A. Antebi A. PLoS Genet. 2007 Sep;3(9):1565-71. doi: 10.1371/journal.pgen.0030129. PLoS Genet. 2007. PMID: 17907808 Free PMC article. Review. - C. elegans SWAN-1 Binds to EGL-9 and regulates HIF-1-mediated resistance to the bacterial pathogen Pseudomonas aeruginosa PAO1.
Shao Z, Zhang Y, Ye Q, Saldanha JN, Powell-Coffman JA. Shao Z, et al. PLoS Pathog. 2010 Aug 26;6(8):e1001075. doi: 10.1371/journal.ppat.1001075. PLoS Pathog. 2010. PMID: 20865124 Free PMC article.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
- Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans . Curr Opin Immunol. 2005;17:4–10. - PubMed
- Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. - PubMed
- Gravato-Nobre MJ, Hodgkin J. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell Microbiol. 2005;7:741–751. - PubMed
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK040561/DK/NIDDK NIH HHS/United States
- R01 AG024425/AG/NIA NIH HHS/United States
- K08 AI053595/AI/NIAID NIH HHS/United States
- AI064332/AI/NIAID NIH HHS/United States
- R56 AG024425/AG/NIA NIH HHS/United States
- AG024425-01/AG/NIA NIH HHS/United States
- P30 DK040561-11/DK/NIDDK NIH HHS/United States
- AI053595/AI/NIAID NIH HHS/United States
- R01 AI064332/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous