Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast - PubMed (original) (raw)
Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast
Rachel Kama et al. Mol Cell Biol. 2007 Jan.
Abstract
BTN2 gene expression in the yeast Saccharomyces cerevisiae is up-regulated in response to the deletion of BTN1, which encodes the ortholog of a human Batten disease protein. We isolated Btn2 as a Snc1 v-SNARE binding protein using the two-hybrid assay and examined its role in intracellular protein trafficking. We show that Btn2 is an ortholog of the Drosophila and mammalian Hook1 proteins that interact with SNAREs, cargo proteins, and coat components involved in endosome-Golgi protein sorting. By immunoprecipitation, it was found that Btn2 bound the yeast endocytic SNARE complex (e.g., Snc1 and Snc2 [Snc1/2], Tlg1, Tlg2, and Vti1), the Snx4 sorting nexin, and retromer (e.g., Vps26 and Vps35). In in vitro binding assays, recombinant His(6)-tagged Btn2 bound glutathione S-transferase (GST)-Snc1 and GST-Vps26. Btn2-green fluorescent protein and Btn2-red fluorescent protein colocalize with Tlg2, Snx4, and Vps27 to a compartment adjacent to the vacuole that corresponds to a late endosome. The deletion of BTN2 blocks Yif1 retrieval back to the Golgi apparatus, while the localization of Ste2, Fur4, Snc1, Vps10, carboxypeptidases Y (CPY) and S (CPS), Sed5, and Sec7 is unaltered in btn2Delta cells. Yif1 delivery to the vacuole was observed in other late endosome-Golgi trafficking mutants, including ypt6Delta, snx4Delta, and vps26Delta cells. Thus, Btn2 facilitates specific protein retrieval from a late endosome to the Golgi apparatus, a process which may be adversely affected in patients with Batten disease.
Figures
FIG. 1.
BTN2 encodes a Hook1 ortholog as shown by MUSCLE alignments of Btn2 from Saccharomyces cerevisiae (sc Btn2; GenBank accession no. NP_011658.1), Drosophila Hook1 (dm Hook1; GenBank accession no. NP_476573.1), and human Hook1 (hs Hook1; GenBank accession no. NP_056972.1). Amino acid identities shared between two or more alignments are shown in black boxes, while similarities are shown in light (less similar) and dark (more similar) gray boxes. Numbers correspond to the amino acid, while periods indicate gaps introduced into the sequence by the program.
FIG. 2.
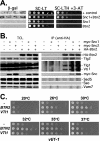
Btn2 interacts with Snc1 and the yeast endocytic SNARE complex. (A) Snc1 interacts with Btn2, as assayed using the two-hybrid β-galactosidase and 3-AT growth assays. Yeast (Y153 cells) were transformed with plasmids expressing the Gal4 DNA binding domain fused to Snc1 (pAS1-Snc1Δ) and full-length Btn2 fused to the Gal4 transactivating domain (pACT-Btn2), together or alone. Transformants were either grown as patches on plates and then replica plated onto nitrocellulose fibers (left panel) or grown to mid-log phase in liquid culture prior to serial dilution and plating by drops onto solid medium (middle and right panels). In addition, positive control (+control) cells expressing Rb and PP1 from plasmids (pAS/N-RB and pPP1α) were used in parallel. For the β-galactosidase (β-gal) assay, cells replica plated onto filters were lysed in liquid nitrogen and measured for β-galactosidase activity using standard techniques. For assaying resistance to 3-aminotriazole (3-AT) and growth in the absence of histidine, cells were plated onto either control selective medium (SC-LT) or medium lacking histidine and containing 25 mM 3-AT (SC-HLT +3-AT). Cells were grown for 60 h at 26°C. (B) The yeast endocytic SNARE complex coimmunoprecipitates with Btn2. Wild-type yeast (W303-1a) expressing HA-Btn2 from a multicopy plasmid (pAD54-Btn2) were transformed with single-copy plasmids expressing myc-Snc1 (pHADH-myc-cSnc1) or myc-Snc2 (pHADH-myc-Snc2), or empty vector alone (pAD11) as control. In addition, wild-type cells bearing a control multicopy plasmid (pAD54) were transformed with the same single-copy plasmids expressing myc-Snc1 or myc-Snc2 or an empty vector as controls. Cells were grown to mid-log phase and processed for immunoprecipitation with anti-HA antibodies. Precipitates (IP) formed from each reaction mixture (500 μg total protein per reaction) and TCLs (20 μg protein per lane) were resolved with SDS-PAGE and detected by Western blotting with a variety of antisera. Shown are blots detected with anti-HA antibody (1:7,000), which reveals that HA-Btn2 is expressed as a doublet. Also shown are blots detected with antisera to Tlg2 (1:1,000), Vti1 and Tlg1 (added together, 1:2,000 and 1:3,000, respectively), myc epitope (to detect myc-Snc1 or myc-Snc2, 1:1,000), Sed5 (1:3,000), Sso1/2 (1:3,000), and Vam7 (1:5,000). Based upon densitometric analysis of the individual Tlg1, Tlg2, and Vti1 signals in the three HA-Btn2-containing IP lanes, we estimate that an average of ∼4% of the endocytic SNARE complex coimmunoprecipitated with HA-Btn2 in the reaction mixtures after normalization for Btn2 precipitation (immunoprecipitated 3.8% Vti1, 3.1% Tlg1, and 4.2% Tlg2; data not shown). (C) Overexpression of BTN2 enhances the growth of vti1-1 cells. vti1-1 cells bearing a control vector (pAD54) or vectors overexpressing HA-BTN2 or HA-VTI1 (pAD54-BTN2 or pAD54-VTI1, respectively) were grown to mid-log phase at 26°C before serial dilution (10×) and plating by drops onto prewarmed solid medium. Plates were grown for 2 to 3 days at the indicated temperatures before photodocumentation.
FIG. 3.
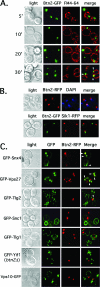
Btn2resides in a late endocytic compartment and colocalizes in parts with Snx4, Tlg2, and Vps27 (arrowheads). (A) Btn2-GFP resides in a late endocytic compartment. Wild-type cells (W303-1b) expressing Btn2-GFP from a single-copy plasmid (pRS316-myc-BTN2-GFP) were grown to mid-log phase on selective synthetic medium at 26°C. Cells were either pulse-labeled with FM4-64 (5.4 μM final concentration) on ice (45 min) and then chased for varying amounts of time (5 to 20 min) at 26°C or were pulse-labeled and chased (30 min) at 26°C. Elapsed chase times are shown in minutes (′). All labeling was performed in the dark. Merge indicates the merger of the GFP and FM4-64 fluorescence windows. (B) Btn2-GFP does not localize to the nucleus. Top panels: wild-type cells (W303-1b) expressing Btn2-RFP from a single-copy plasmid (pRS316-myc-BTN2-mRFP) were grown to mid-log phase on selective synthetic medium at 26°C, labeled briefly with Hoechst dye (50 μg/ml Hoechst 33342 for 10 min at room temperature; Molecular Probes), and visualized by confocal microscopy. Bottom panels: SIK1-RFP cells (ATCC 201389) were transformed with a single-copy plasmid (pRS316-myc-BTN2-GFP) expressing Btn2-GFP. Cells were grown to mid-log phase at 26°C and visualized by confocal microscopy. Merge indicates the merger of GFP and RFP or RFP and DAPI fluorescence windows. (C) Btn2-RFP colocalizes in part with the Snx4, Tlg2, and Vps27 endosomal markers (arrowheads). Wild-type cells (W303-1b) expressing Btn2-RFP from a single-copy plasmid (pRS313-myc-BTN2-mRFP) were transformed with plasmids expressing GFP-tagged forms of Snx4 (pAD54-GFP-SNX4), Vps27 (pGO426-GFP-VPS27), Tlg2 (pRS315-GFP-TLG2), Snc1 (pADHU-GFP-cSNC1), Tlg1 (pRS315-GFP-TLG1), Vps10 (pAD54-VPS10-GFP), and Yif1 (pRS316-GFP-YIF1) (see Table 2 for plasmid details). Cells were grown to mid-log phase at 26°C on selective synthetic medium prior to visualization by confocal microscopy. btn2Δ cells (RKY4) were used to coexpress GFP-Yif1 (pRS316-GFP-YIF1) and Btn2-RFP (pRS313-myc-BTN2-mRFP), instead of wild-type cells. Merge indicates the merger of the GFP and RFP fluorescence windows.
FIG. 4.
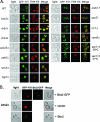
Btn2 localization is not altered in cells deficient in endocytosis and endosomal protein sorting. (A) Btn2 localization is not altered in cells deficient in endocytosis and endosomal protein sorting. A single-copy plasmid (pRS316-myc-BTN2-GFP) expressing Btn2-GFP was transformed into wild-type (WT) cells (W303-1b) or a variety of yeast mutants. The latter includes strains bearing deletions of RCY1, CHS4, GCS1, RHB1 (both Euroscarf and JU28-1 rhb1Δ strains tested; Euroscarf strain shown), SNC1, SNC1/2 (sncΔ; JG8 T15:85), VPS23, VPS27, or VPS28, as well as strains bearing temperature-sensitive mutations in genes SEC27 (RSY1312), SEC33 (RDY260), and SEC21 (RSY1309) COPI subunits or in END4 (RH268-1c). Cells were grown to mid-log phase at 26°C (permissive temperatures) on selective synthetic medium prior to visualization by confocal microscopy. (B) The colocalization between Btn2 and Snx4 or Vps27 is not altered in vps27Δ and snx4Δ cells, respectively. vps27Δ and snx4Δ cells expressing Btn2-RFP from a single-copy plasmid (pRS313-myc-BTN2-mRFP) were transformed with plasmids expressing either GFP-Snx4 (pAD54-GFP-SNX4) or GFP-Vps27 (pGO426-GFP-VPS27), respectively. Merge indicates the merger of the GFP and RFP fluorescence windows. (C) Btn2-GFP(x2) expressed from the genomic BTN2 locus labels multiple large compartments. Yeast bearing BTN2 tagged with GFP(x2) at its genomic locus (RKY7 cells) were grown to mid-log phase at 26°C prior to visualization by confocal microscopy. Merge indicates the merger between the light microscopy and GFP fluorescence windows.
FIG. 5.
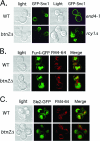
Yif1 is mislocalized to the vacuole in btn2Δ, rhb1Δ, vps26Δ, snx4Δ, chs4Δ, tlg2Δ, and ypt6Δ cells. (A) GFP-Yif1 is mislocalized to the vacuole in btn2Δ, chs4Δ, rhb1Δ, snx4Δ, tlg2Δ, vps26Δ, and ypt6Δ cells. A single-copy plasmid expressing GFP-Yif1 (pRS316-GFP-YIF1) was transformed into wild-type (WT) cells (both Euroscarf and W303-1b strains tested; Euroscarf cells shown) or cells bearing mutations of BTN2 (both Euroscarf and RKY4 _btn2_Δ strains tested; Euroscarf cells shown), RHB1 (both Euroscarf and JU28-1 rhb1Δ strains examined; Euroscarf strain shown), CHS4, VPS26, YPT6, SNX4, TLG2 VAM6, END4 (RH268-1c), RIC1, VPS51, and SEC7 (RSY979). Cells were grown to mid-log phase at 26°C on selective synthetic medium prior to pulse-labeling with FM4-64 (5.4 μM final concentration; 30 min at 26°C) and chase (30 min at 26°C) before visualization using confocal microscopy. sec7-5 (RSY979) cells were grown to mid-log phase at 26°C (permissive temperature) and either maintained and labeled with FM4-64 at 26°C or shifted to 37°C for 30 min prior to pulse-labeling with FM4-64 (30 min; 37°C) and chase (30 min; 37°C) before visualization. Note labeling of the vacuole by GFP in btn2Δ, chs4Δ, rhb1Δ, snx4Δ, tlg2Δ, vps26Δ, and ypt6Δ cells. See Table 3 for statistics regarding either Golgi or vacuolar GFP-Yif1 localization in the various strains examined. Merge indicates the merger of the GFP and FM4-64 fluorescence windows. (B) Btn2 and Btn2-GFP restore RFP-Yif1 localization to btn2Δ cells. btn2Δ cells expressing RFP-Yif1 from a multicopy plasmid (pAD54-RFP-YIF1) were transformed with either a control plasmid (pRS313) or single-copy plasmids expressing Btn2 or Btn2-GFP (pRS313-BTN2 or pRS313-BTN2-GFP). Cells were grown to mid-log phase at 26°C prior to visualization. Merge indicates the merger of light and fluorescence microscopy images.
FIG. 6.
The localization of GFP-tagged Snc1, Fur4, and Ste2 is unaltered in btn2Δ cells. (A) GFP-Snc1 localization is unaltered in btn2Δ cells. Wild-type (Euroscarf and W303-1b strains tested; Euroscarf cells shown), btn2Δ (Euroscarf and RKY5 btn2Δ strains examined; Euroscarf shown), end4-1, and rcy1Δ cells expressing GFP-Snc1 from a single-copy plasmid (pRS315-GFP-cSNC1) were grown to mid-log phase on synthetic selective medium at 26°C prior to visualization by confocal microscopy. (B) Fur4-GFP localization is unaltered in cells lacking BTN2. Wild-type (Euroscarf and W303-1b strains tested; Euroscarf cells shown) and btn2Δ (Euroscarf and RKY4 btn2Δ strains examined; Euroscarf cells shown) cells expressing Fur4-GFP from a single-copy plasmid (pRS316-FUR4-GFP) were grown to mid-log phase on synthetic selective medium at 26°C prior to pulse-labeling with FM4-64 (5.4 μM final concentration; 30 min at 26°C) and chase (30 min at 26°C) before visualization using confocal microscopy. (C) Ste2-GFP localization is unaltered in cells lacking BTN2. Wild-type (W303-1b) and btn2Δ (RKY5 btn2Δ strain examined) cells expressing Ste2-GFP from a single-copy plasmid (pRS314-STE2-GFP) were grown to mid-log phase on synthetic selective medium at 26°C prior to pulse-labeling with FM4-64 (5.4 μM final concentration; 30 min at 26°C) and chase (30 min at 26°C), before visualization using confocal microscopy. Merge indicates the merger of GFP and FM4-64 (B and C) fluorescence microscopy images.
FIG. 7.
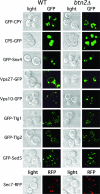
The deletion of BTN2 does not affect the trafficking of a wide variety of other endosomal cargo proteins. Wild-type (WT) and btn2Δ cells expressing CPY-GFP [pGALΔ BglII-CPY(1-50)GFP], CPS-GFP (pGO426-CPS1-GFP), GFP-Snx4 (pAD54-GFP-SNX4), GFP-Vps27 (pGO426-GFP-VPS27), Vps10-GFP (pAD54-VPS10-GFP), GFP-Tlg1 (pRS315-GFP-TLG1), GFP-Tlg2 (pRS315-GFP-TLG2), GFP-Sed5 (pRS315-GFP-SED5), or Sec7-RFP were grown to mid-log phase on synthetic selective medium at 26°C prior to visualization by confocal microscopy. Protein expression was performed using the Euroscarf wild-type and btn2Δ cells, except in the case of CPS-GFP expression, which was performed with W303-1b and RKY4 btn2Δ cells, and Sec7-RFP expression, which was performed using the W303-1a-SEC7-RFP and RKY6 strains.
FIG. 8.
Btn2 interacts with a complex containing Yif1, Snx4, and retromer. (A) GFP-Yif1 is more degraded in cells lacking BTN2. Wild-type (WT) (W303-1b) and btn2Δ (RKY4) cells expressing GFP-Yif1 (GFP-Yif1) from a single-copy plasmid (pRS316-GFP-YIF1) were grown to mid-log phase and lysed. Equal amounts of protein from TCLs were subjected to SDS-PAGE and Western analysis with antibodies against GFP (1:1,000; left panel) to detect both GFP and GFP-Yif1, and intermediates. In the right panels, antibodies against CPY (1:1,000), Mnn1 (1:1,000), Dpm1 (1:1,000), Sec22 (1:1,000), Bet1 (1:1,000), and actin (1:10,000) were used. (B) Btn2 coimmunoprecipitates with Vps26 and Vps35. Wild-type, vps17Δ, and vps26Δ cells (all Euroscarf strains) expressing myc-Btn2-RFP from a single-copy plasmid (+) (pRS316-myc-Btn2-mRFP) or bearing a control vector (−) (pRS316) were grown to mid-log phase and processed for immunoprecipitation. Samples from the TCLs and precipitates (IP) were resolved by SDS-PAGE and detected in blots with anti-Vps26 (1:1,000), anti-Vps35 (1:1,000), and anti-myc (1:1,000) antibodies. (C) Btn2 coimmunoprecipitates with Snx4. Wild-type yeast (BY4741) expressing myc-Btn2 from a single copy plasmid (+) (pRS316-myc-Btn2-mRFP) or bearing a control vector (−) (pRS316) were transformed with plasmids expressing GFP-Snx4 (pAD54-GFP-SNX4**), GFP-Yif1 (pAD54-GFP-YIF1), GFP-Snc1 (pAD54-GFP-cSNC1), or a control vector (−) (pAD54)**. Cells were grown to mid-log phase and processed for immunoprecipitation. Samples from the TCLs and precipitates were resolved by SDS-PAGE and detected in blots with anti-GFP (1:1,000) and anti-myc (1:1,000) antibodies. (D) His6-Btn2 binds directly to GST-Snc1 and GST-Vps26. Pull-down experiments (left panels) were performed by mixing equal amounts of protein (10 μg) of purified recombinant His6-Btn2 and GST-tagged Snx4, Vps17, Vps26, Tlg2, Snc1, and Sso1, and GST alone. After binding, nickel-charged beads were used to precipitate complexes which were resolved by SDS-PAGE and detected by anti-GST (1:500) or anti-His6 (1:1,000) antibodies. Arrowheads indicate the detection of GST-Vps26 and GST-Snc1 in the blots. Samples (10 μg) of the purified recombinant proteins (input; right panels) were also resolved by SDS-PAGE and detected with anti-GST (1:500) or anti-His6 (1:1,000) antibodies. Arrowheads indicate the appropriate full-length recombinant proteins.
FIG. 9.
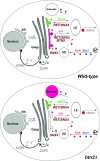
A model for Btn2 function in late endosome-Golgi protein sorting. Wild-type cells: the Snc1 exo/endocytic v-SNARE (blue) and Fur4 (red) are both endocytosed and delivered by endocytic vesicles to the early endosome (EE). Snc1 is delivered to the trans Golgi location in a manner dependent upon Snx4, Rcy1, Ric1, and Ypt31/32, etc. (later components are not listed). Fur4 is trafficked to the late endosome (LE) for either recycling to the plasma membrane or delivery to the vacuole for degradation. Retrieval to the plasma membrane is presumably mediated by high-density secretory vesicles (HDSVs), which originate from late endosomes (35, 37). A Golgi marker, Yif1 (purple), exits the Golgi apparatus to the late endosome but is retrieved in a retromer-, Snx4-, and Btn2-dependent fashion back to the Golgi apparatus. Vps10 (green), the CPY receptor, is also recycled to the Golgi apparatus from a late endosome via retromer and Snx4 but in a manner independent of Btn2. btn2Δ cells: in the absence of BTN2, the recycling of Yif1 (but not the other tested Golgi markers, e.g., Sed5 and Sec7) to the Golgi apparatus is blocked and the protein is sent to the vacuole for degradation. The endosomal sorting of Vps10, Snc1, and Fur4 is unaffected in the absence of Btn2. Thus, we predict that Btn2 acts, along with retromer and Snx4, to mediate the retrieval of Yif1 from a sorting late endosome back to the Golgi apparatus.
Similar articles
- Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast.
Kanneganti V, Kama R, Gerst JE. Kanneganti V, et al. Mol Biol Cell. 2011 May 15;22(10):1648-63. doi: 10.1091/mbc.E10-11-0878. Epub 2011 Mar 25. Mol Biol Cell. 2011. PMID: 21441304 Free PMC article. - Btn3 regulates the endosomal sorting function of the yeast Ent3 epsin, an adaptor for SNARE proteins.
Morvan J, de Craene JO, Rinaldi B, Addis V, Misslin C, Friant S. Morvan J, et al. J Cell Sci. 2015 Feb 15;128(4):706-16. doi: 10.1242/jcs.159699. Epub 2014 Dec 15. J Cell Sci. 2015. PMID: 25512335 - The yeast Batten disease orthologue Btn1 controls endosome-Golgi retrograde transport via SNARE assembly.
Kama R, Kanneganti V, Ungermann C, Gerst JE. Kama R, et al. J Cell Biol. 2011 Oct 17;195(2):203-15. doi: 10.1083/jcb.201102115. Epub 2011 Oct 10. J Cell Biol. 2011. PMID: 21987636 Free PMC article. - SNARE Protein Snc1 Is Essential for Vesicle Trafficking, Membrane Fusion and Protein Secretion in Fungi.
Adnan M, Islam W, Waheed A, Hussain Q, Shen L, Wang J, Liu G. Adnan M, et al. Cells. 2023 Jun 5;12(11):1547. doi: 10.3390/cells12111547. Cells. 2023. PMID: 37296667 Free PMC article. Review. - Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae.
Bowers K, Stevens TH. Bowers K, et al. Biochim Biophys Acta. 2005 Jul 10;1744(3):438-54. doi: 10.1016/j.bbamcr.2005.04.004. Biochim Biophys Acta. 2005. PMID: 15913810 Review.
Cited by
- Molecular mechanisms of spatial protein quality control.
Alberti S. Alberti S. Prion. 2012 Nov-Dec;6(5):437-42. doi: 10.4161/pri.22470. Epub 2012 Oct 10. Prion. 2012. PMID: 23051707 Free PMC article. - Curing of the [URE3] prion by Btn2p, a Batten disease-related protein.
Kryndushkin DS, Shewmaker F, Wickner RB. Kryndushkin DS, et al. EMBO J. 2008 Oct 22;27(20):2725-35. doi: 10.1038/emboj.2008.198. Epub 2008 Oct 2. EMBO J. 2008. PMID: 18833194 Free PMC article. - How Do Yeast Cells Contend with Prions?
Wickner RB, Edskes HK, Son M, Wu S, Niznikiewicz M. Wickner RB, et al. Int J Mol Sci. 2020 Jul 3;21(13):4742. doi: 10.3390/ijms21134742. Int J Mol Sci. 2020. PMID: 32635197 Free PMC article. Review. - Transcriptional profiling of Zygosaccharomyces bailii early response to acetic acid or copper stress mediated by ZbHaa1.
Antunes M, Palma M, Sá-Correia I. Antunes M, et al. Sci Rep. 2018 Sep 20;8(1):14122. doi: 10.1038/s41598-018-32266-9. Sci Rep. 2018. PMID: 30237501 Free PMC article. - Hook2 is involved in the morphogenesis of the primary cilium.
Baron Gaillard CL, Pallesi-Pocachard E, Massey-Harroche D, Richard F, Arsanto JP, Chauvin JP, Lecine P, Krämer H, Borg JP, Le Bivic A. Baron Gaillard CL, et al. Mol Biol Cell. 2011 Dec;22(23):4549-62. doi: 10.1091/mbc.E11-05-0405. Epub 2011 Oct 12. Mol Biol Cell. 2011. PMID: 21998199 Free PMC article.
References
- Abeliovich, H., E. Grote, P. Novick, and S. Ferro-Novick. 1998. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol. Chem. 273:11719-11727. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous