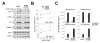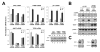Acetylation of the p53 DNA-binding domain regulates apoptosis induction - PubMed (original) (raw)
Acetylation of the p53 DNA-binding domain regulates apoptosis induction
Stephen M Sykes et al. Mol Cell. 2006.
Abstract
The ability of p53 to induce apoptosis plays an important role in tumor suppression. Here, we describe a previously unknown posttranslational modification of the DNA-binding domain of p53. This modification, acetylation of lysine 120 (K120), occurs rapidly after DNA damage and is catalyzed by the MYST family acetyltransferases hMOF and TIP60. Mutation of K120 to arginine, as occurs in human cancer, debilitates K120 acetylation and diminishes p53-mediated apoptosis without affecting cell-cycle arrest. The K120R mutation selectively blocks the transcription of proapoptotic target genes such as BAX and PUMA while the nonapoptotic targets p21 and hMDM2 remain unaffected. Consistent with this, depletion of hMOF and/or TIP60 inhibits the ability of p53 to activate BAX and PUMA transcription. Furthermore, the acetyllysine 120 (acetyl-K120) form of p53 specifically accumulates at proapoptotic target genes. These data suggest that K120 acetylation may help distinguish the cell-cycle arrest and apoptotic functions of p53.
Figures
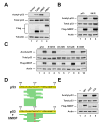
Figure 1. hMOF and TIP60 enhance acetylation of p53
(A) H1299 cells were co-transfected with p53 and either vector, FLAG-TIP60, FLAG-hMOF, or FLAG-HBO1. Following 4 hours of treatment with 10mM sodium butyrate, cells were lysed and subjected to immunoprecipitation (IP) with a pan-acetyl-lysine (AcK) antibody. Precipitates were blotted with a p53 antibody to quantify p53 acetylation. Input lysates were also blotted with FLAG, p53, and tubulin antibodies. (B) H1299 cells were co-transfected with either wild type or 9K/R in the presence or absence of a plasmid expressing FLAG-hMOF. Acetylation was assessed by IP using the strategy described in (A). Total p53 expression was assessed by western blot of input lysates. (C) Expression vectors for mutants of p53 carrying individual lysine-to-arginine substitution mutations within the DNA-binding domain of p53 were transfected into H1299 cells with or without a plasmid expressing FLAG-hMOF. Acetylation was assessed by IP using the strategy described above. Total p53 expression was assessed by western blot of input lysates. (D) FLAG epitope-tagged p53 expressed in either the presence or the absence of hMOF was affinity purified from transiently transfected H1299 cells. The purified p53 proteins were subjected to μLC-MS/MS analysis to identify in vivo sites of hMOF-mediated acetylation. The sequence of the FLAG-epitope tagged p53 protein is displayed with green bars indicating the position of the tryptic or chymotryptic peptide spectra detected in the μLC-MS/MS analysis from the sample co-expressed with hMOF. Acetylated lysines are displayed in red. (E) H1299 cells were co-transfected with p53 and plasmids expressing FLAG-hMOF, FLAG-hMOF HAT (represents a 60-amino acid deletion within the HAT domain of hMOF), or FLAG-hMOF CD (represents a 66-amino acid deletion of the hMOF chromodomain). Following transfection cells were lysed and subjected to IP with a pan-acetyl-lysine antibody. To assess p53 acetylation pan-acetyl-precipitates were subjected to western blot with a p53 antibody (Top panel). Input lysates were also analyzed by western blot with the indicated antibodies (bottom three panels).
Figure 2. hMOF directly acetylates p53 in vitro
(A) Increasing amounts of recombinant full-length hMOF and radioactive acetyl Co-A were incubated with and without recombinant human p53 (amino acid residues 94 to 312). After incubation, liquid scintillation was used to assess enzymatic activity. Increasing amounts of full-length hMOF were incubated with a histone H4 peptide (S1-R19) as a positive control. (B) Both wild type and K120R versions of recombinant human p53 (described above) were purified from bacterial extracts. Subsequently each p53 protein was incubated separately with varying concentrations of hMOF. After incubation, liquid scintillation counting was used to detect the acetylation of wild-type and mutant p53.
Figure 3. Acetylation of lysine 120 catalyzed by hMOF and TIP60 increases in response to genotoxic stress
(A) H1299 cells were transfected with p53 in the presence or absence of hMOF. After transfection, cells were treated with 10mM sodium butyrate for 2–4 hours. Following treatment, the cell lysate was divided into two aliquots. One aliquot was subjected to immunoprecipitation (IP) with AcK120 antibody pre-blocked with 250μM modified peptide. The other aliquot was subjected to IP with AcK120 antibody pre-blocked with 25μM of unmodified peptide. Precipitates were then western blotted with a p53 antibody to quantify lysine-120 acetylation. Western blots were performed on the input lysates to ensure that equal amounts of p53 were used for each IP. (B) H1299 cells were transfected with wild-type p53 in the presence or absence of hMOF. In parallel, H1299 cells were co-transfected with p53-(K120R) and either vector control or hMOF. After transfection, cells were treated with 10mM sodium butyrate for 2–4 hours. Following treatment, cells were lysed and subjected to IP with an AcK120 antibody. Precipitates were western blotted with a p53 antibody to quantify lysine-120 acetylation. Input lysates were also subjected to western blot as described above. (C) U2OS cells were treated with 25 J/m2 UV, 5μM camptothecin (CPT), 10 Gy γ-irradiation (γIR), or 0.5 μg/ml adriamycin (Adr) and then harvested at the indicated times. Ninety minutes prior to each time point, cells were treated with 10mM Sodium Butyrate. K120 acetylation was assessed as described above. (D) U2OS cells were infected with recombinant lentiviruses that express shRNA molecules targeted to the corresponding genes. Seventy-two hours following infection, cells from each shRNA condition were treated with 10 Gy γ–Irradiation or left untreated. Two hours after irradiation, cells were treated with 10 mM Sodium Butyrate for 1 hour. Cells were then harvested and subjected to immunoprecipitation and subsequent western blot as described above.
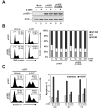
Figure 4. A conservative mutation at lysine 120 specifically reduces p53 mediated-apoptosis
(A) H1299 cells were infected with recombinant retroviruses expressing either p53ER or p53ER-(K120R). Mock infected cells served as a control. Infected cells were lysed and subjected to western blot to assess the expression of each construct. (B) H1299-p53ER and H1299-p53ER-(K120R) cells were treated with increasing concentrations of 4-hydroxytamoxifen (4-OHT) for 24 hours. Following treatment cells were fixed, stained with propidium iodide (PI), and analyzed by FACS to assess cellular DNA content. Left panel displays results obtained from FACS analysis of cells treated with and without 50 nM 4-OHT. Right panel is a graphical representation of FACS data obtained from all treatment conditions. (C) H1299-p53ER and H1299-p53ER-(K120R) cells were treated with 50 nM 4-OHT for 24 hours. Cells were then treated with 5 μM CPT, 0.5 μM Adr, 12 Gy γIR, or 50 J/m2 UV. Twenty-four hours after DNA damage, cells were stained with annexin V-FITC and PI and then analyzed by FACS. Left panel displays plots obtained from FACS analysis of cells treated with CPT. Right panel is a graphical representation of annexin V positive (PI negative) cells collected after the treatment indicated. Error bars represent standard deviation of three independent cell counts.
Figure 5. p53-(K120R) is specifically defective for BAX and PUMA induction
(A) H1299-p53ER and –p53ER-(K120R) cells were treated with 50 or 100 nM 4-OHT or vehicle for 24 hours. Cells were collected, lysed and subjected to western blot with the antibodies indicated. (B) H1299-p53ER and –p53ER-(K120R) cells were treated with 50 or 200nM 4-OHT for 24 hours and subsequently harvested for RNA at 0, 6, 12, 18 and 24 hours. cDNA generated from recovered RNA was amplified with primers to the target genes indicated and quantified by real-time PCR. (C) H1299-p53ER and –p53ER-(K120R) cells were treated with 100nM 4-OHT or vehicle for 6 hours. Cells from each condition were cross-linked and subjected to ChIP analysis with a p53 antibody. Precipitated DNA was recovered and analyzed by real-time PCR with primers that amplify the p53-binding site within the BAX, PUMA, hMDM2, and p21 promoters. Error bars represent standard deviation of three independent reactions.
Figure 6. Depletion of hMOF and TIP60 reduces the ability of p53 to induce BAX and PUMA
(A) MCF-7 cells were infected with recombinant lentiviruses that express shRNA molecules directed against luciferase (control), hMOF, or TIP60. Seventy-two hours following infection, cells were treated with 25 J/m2 UV and then collected 16 hours thereafter. Harvested cells were divided into two aliquots. One aliquot from each treatment condition was harvested for RNA, which was converted to cDNA as described above. cDNA samples were then analyzed by real-time PCR with primers corresponding to the indicated genes. The other aliquot was lysed and subjected to western blot with the antibodies indicated (lower right panel). (B) H1299-p53ER and –p53ER-(K120R) cells were infected with lentiviruses expressing the shRNA molecules indicated. Seventy-two hours following infection, cells were treated with 100 nM 4-OHT or vehicle for 16 hours. Harvested cells were analyzed by western blot with the antibodies indicated. (C) Lentiviral shRNA vectors efficiently reduce protein levels for both hMOF and TIP60. H1299 cells were transfected with either FLAG-hMOF or FLAG-TIP60. Transfected cells were then split into 2 dishes and infected with recombinant lentiviruses expressing either luciferase (LUC), hMOF, or TIP60 shRNA molecules. Seventy-two hours following infection, cells were harvested, lysed and subjected to western blot with the indicated antibodies. Error bars represent standard deviation of three independent reactions.
Figure 7. p53 acetylated at K120 selectively accumulates at pro-apoptotic target gene promoters
(A) LNCaP cells treated with 5 μM CPT for 0, 2, and 6 hours were cross-linked and harvested for ChIP. Lysates from each condition were divided into equal aliquots and incubated with either p53 or AcK120-p53 antibodies. Precipitated DNA was recovered and analyzed by real-time PCR with primers that amplify the area encompassing the p53 binding sites within the promoters indicated. (B) Six hours following 20 J/m2 UV exposure, MCF-7 cells were harvested and analyzed as described in (A). Error bars represent standard deviation of three independent reactions.
Comment in
- To die or not to die: a HAT trick.
Tyteca S, Legube G, Trouche D. Tyteca S, et al. Mol Cell. 2006 Dec 28;24(6):807-8. doi: 10.1016/j.molcel.2006.12.005. Mol Cell. 2006. PMID: 17189182 Review.
Similar articles
- Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis.
Tang Y, Luo J, Zhang W, Gu W. Tang Y, et al. Mol Cell. 2006 Dec 28;24(6):827-39. doi: 10.1016/j.molcel.2006.11.021. Mol Cell. 2006. PMID: 17189186 - ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage.
Liu N, Wang J, Wang J, Wang R, Liu Z, Yu Y, Lu H. Liu N, et al. Cancer Res. 2013 Jun 15;73(12):3749-60. doi: 10.1158/0008-5472.CAN-12-3684. Epub 2013 Apr 10. Cancer Res. 2013. PMID: 23576563 - Phosphorylation of Tip60 by p38α regulates p53-mediated PUMA induction and apoptosis in response to DNA damage.
Xu Y, Liao R, Li N, Xiang R, Sun P. Xu Y, et al. Oncotarget. 2014 Dec 30;5(24):12555-72. doi: 10.18632/oncotarget.2717. Oncotarget. 2014. PMID: 25544752 Free PMC article. - To die or not to die: a HAT trick.
Tyteca S, Legube G, Trouche D. Tyteca S, et al. Mol Cell. 2006 Dec 28;24(6):807-8. doi: 10.1016/j.molcel.2006.12.005. Mol Cell. 2006. PMID: 17189182 Review. - The ARF/oncogene pathway activates p53 acetylation within the DNA binding domain.
Mellert H, Sykes SM, Murphy ME, McMahon SB. Mellert H, et al. Cell Cycle. 2007 Jun 1;6(11):1304-6. doi: 10.4161/cc.6.11.4343. Epub 2007 Jun 24. Cell Cycle. 2007. PMID: 17534149 Review.
Cited by
- Will Sirtuins Be Promising Therapeutic Targets for TBI and Associated Neurodegenerative Diseases?
Yang Q, Zhou Y, Sun Y, Luo Y, Shen Y, Shao A. Yang Q, et al. Front Neurosci. 2020 Jul 31;14:791. doi: 10.3389/fnins.2020.00791. eCollection 2020. Front Neurosci. 2020. PMID: 32848564 Free PMC article. Review. - Alternatively mechanistic insights into acetylation in p53-mediated transcriptional regulation of cancer cell-intrinsic PD-1.
Wen J, Yao H, Cao Z, Wang D. Wen J, et al. Fundam Res. 2022 Apr 1;3(4):647-654. doi: 10.1016/j.fmre.2022.03.012. eCollection 2023 Jul. Fundam Res. 2022. PMID: 38933547 Free PMC article. - H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation.
Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. Lauberth SM, et al. Cell. 2013 Feb 28;152(5):1021-36. doi: 10.1016/j.cell.2013.01.052. Cell. 2013. PMID: 23452851 Free PMC article. - Determinants of p53 DNA binding, gene regulation, and cell fate decisions.
Fischer M, Sammons MA. Fischer M, et al. Cell Death Differ. 2024 Jul;31(7):836-843. doi: 10.1038/s41418-024-01326-1. Epub 2024 Jun 29. Cell Death Differ. 2024. PMID: 38951700 Free PMC article. Review. - Tumor suppressor p53: from engaging DNA to target gene regulation.
Sammons MA, Nguyen TT, McDade SS, Fischer M. Sammons MA, et al. Nucleic Acids Res. 2020 Sep 18;48(16):8848-8869. doi: 10.1093/nar/gkaa666. Nucleic Acids Res. 2020. PMID: 32797160 Free PMC article.
References
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. - PubMed
- Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. - PubMed
- Chipuk JE, Maurer U, Green DR, Schuler M. Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell. 2003;4:371–381. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA090465/CA/NCI NIH HHS/United States
- R01 CA098172/CA/NCI NIH HHS/United States
- R01 CA090465/CA/NCI NIH HHS/United States
- R01 GM060293/GM/NIGMS NIH HHS/United States
- CA098172/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous