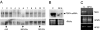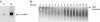RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex - PubMed (original) (raw)
RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex
Rustem T Omarov et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Tomato bushy stunt virus (TBSV) and other tombusviruses encode a p19 protein (P19), which is a suppressor of RNAi. Wild-type TBSV or p19-defective mutants initially show a similar infection course in Nicotiana benthamiana, but the absence of an active P19 results in viral RNA degradation followed by recovery from infection. P19 homodimers sequester 21-nt virus-derived duplex siRNAs, and it is thought that this prevents the programming of an antiviral RNA-induced silencing complex to avoid viral RNA degradation. Here we report on chromatographic fractionation (gel filtration, ion exchange, and hydroxyapatite) of extracts from healthy or infected Nicotiana benthamiana plants in combination with in vitro assays for ribonuclease activity and detection of TBSV-derived siRNAs. Only extracts of plants infected with p19 mutants provided a source of sequence-nonspecific but ssRNA-targeted in vitro ribonuclease activity that coeluted with components of a wide molecular weight range. In addition, we isolated a discrete approximately 500-kDa protein complex that contained approximately 21-nt TBSV-derived siRNAs and that exhibited ribonuclease activity that was TBSV sequence-preferential, ssRNA-specific, divalent cation-dependent, and insensitive to a ribonuclease inhibitor. We believe that this study provides biochemical evidence for a virus-host system that infection in the absence of a fully active RNAi suppressor induces ssRNA-specific ribonuclease activity, including that conferred by a RNA-induced silencing complex, which is likely the cause for the recovery of plants from infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
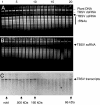
In vitro RNA cleavage assays with gel-filtration fractions collected from healthy or wt TBSV-infected plants. (A and B) Healthy (A) or TBSV-infected (B) N. benthamiana extracts were subjected to gel-filtration chromatography on a Sephacryl S-200 HR column, and collected fractions were tested for the ability to cleave total RNA extracted from TBSV-infected N. benthamiana plants, as visualized by ethidium bromide staining of nucleic acids. (C) Fractions shown in B were tested for in vitro cleavage of TBSV transcripts visualized by Northern blot hybridization. Select numbers on the top indicate the order of collected fractions (–20). Arrows and abbreviations to the right indicate the positions of plant chromosomal DNA (plant DNA), viral dsRNA (TBSV dsRNA) or viral ssRNA (TBSV ssRNA), and plant rRNAs. Arrows on the bottom indicate the fractions representing the void volume and the extrapolated elution profile of standard MW markers on the same column.
Fig. 2.
In vitro RNA cleavage assays with fractions of plants infected with a TBSV mutant that does not express P19. (A) Gel-filtration chromatography fractions were collected from N. benthamiana infected with TBSV-dP19, as for Fig. 1, and tested for cleavage of total RNA extracted from wt TBSV-infected N. benthamiana plants. (B) The same fractions were tested for cleavage of TBSV transcripts that were synthesized and tested in the presence of RNase A inhibitor. The asterisk indicates a pair of closely spaced bands, the upper band corresponding to TBSV genomic ssRNA and the lower band corresponding to TBSV dsRNA of subgenomic RNA2 (18), which resisted degradation. Visualization and labeling is as for Fig. 1.
Fig. 3.
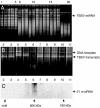
In vitro RNA cleavage assays and siRNA detection in fractions from plants infected with a TBSV mutant expressing P19/R43W. (A and B) GFs were tested (as Fig. 1) for cleavage of total RNA extracted from TBSV-infected N. benthamiana plants (A) or in vitro generated TBSV transcripts (the higher band represents the DNA template) (B). Symbols, numbers, and abbreviations are as for Fig. 1. (C) Fractions obtained after gel filtration were concentrated and subjected to urea-gel denaturing electrophoresis followed by Northern blot hybridization by using a TBSV-specific probe. The arrow on the right indicates the position of TBSV ≈21-nt siRNAs.
Fig. 4.
Activity tests and siRNA detection for ion-exchange fractions obtained from N. benthamiana plants infected with TBSV-dP19. (A) Denaturing gel-electrophoresis followed by Northern blot hybridization for the detection of ≈21-nt TBSV-specific siRNAs in fractions collected on DEAE ion-exchange chromatography (every other fraction was tested). The positions of RNA size markers are indicated on the rightmost side of the blot. (B) Native agarose gel electrophoresis followed by Northern blot hybridization to detect TBSV-specific siRNAs. (C and D) Fractions were tested for in vitro cleavage of TBSV transcripts followed by Northern blot hybridization (C) or total RNA extracted from TBSV-infected N. benthamiana plants as visualized by ethidium bromide staining after treatments (D). Numbers on the top indicate the individually collected fractions (–37). Arrows on the right indicate positions of TBSV RNA (for this particular experiment, the dsRNA is not visible on the gel). Numbers on the bottom indicate the increasing NaCl concentration.
Fig. 5.
TBSV-specific ribonuclease activity upon ion-exchange chromatography followed by gel filtration. Fractions containing ≈21-nt TBSV siRNAs after DEAE ion-exchange chromatography with extracts from N. benthamiana plants infected with TBSV-dP19 were concentrated and fractionated on a Sephacryl S-200 HR column. (A) Aliquots of eluted fractions were tested for RNA cleavage with TBSV in vitro synthesized transcripts (Upper) or with TBSV gRNA extracted from infected plants (Lower), detected by Northern blot hybridization with a TBSV-specific probe. (B) Aliquots of GF-6 (5 μl) were incubated with total RNA extracted from TBSV-infected plants. Also shown is the control (C) incubation of RNA with elution buffer. TBSV gRNA was detected by Northern blot hybridization with a TBSV-specific probe (Upper), and the ethidium bromide stained gel (Lower) shows the plant rRNAs in the same samples. (C) Agarose gel electrophoreses of transcripts from TBSV, satellite panicum mosaic virus (SPMV), or HFi22 (representing a host mRNA), after incubation with GF-6 or an inactive control (C) fraction. Visualization is by ethidium bromide staining.
Fig. 6.
Isolation of an siRNA-containing ribonuclease complex by using a hydroxyapatite column followed by ion-exchange chromatography. (A) Hydroxyapetite fractions with ribonuclease activity were combined and subjected to ion-exchange chromatography. The active fractions were again pooled and subjected to RNA isolation followed by gel electrophoresis and Northern blot hybridization with a TBSV-specific probe (F*). The right lane was loaded with a ≈21 TBSV siRNA as a positive marker (M). (B) A time course (minutes) RNA cleavage test with the siRNA-containing ribonuclease fraction from A (F*) mixed with in vitro generated TBSV transcripts, detected by Northern blot hybridization. The reactions were stopped by the addition of buffer (100 mM Tris·HCl, pH 7.4/20 mM sodium EDTA). The arrow on the right indicates the position of full-length TBSV transcripts; degradation products, some of seemingly distinct sizes, migrate underneath.
Fig. 7.
Effects of divalent metal cations and substrate concentration on the high-MW ribonuclease activity. (A) A fraction similarly prepared as described for Fig. 6 was tested for ribonuclease activity toward TBSV transcripts in the presence of water, 2 mM MnCl2, 2 mM MgCl2, or 10 mM EDTA. A control (C) treatment lacked nuclease addition to transcripts. The treatments are indicated at the top of each lane, and the RNA is detected by Northern blot hybridization. The arrow on the right indicates the position of TBSV RNA transcripts. (B) Ribonuclease activity was tested with increased concentrations of total RNA extracted from TBSV-infected plants (indicated on top). (Upper) Visualization by ethidium bromide staining of the gel before transfer for the Northern blot hybridization shows the chromosomal DNA, TBSV dsRNA and ssRNA, and rRNAs, as indicated on the right. (Lower) The effect of ribonuclease addition on the integrity of TBSV gRNA as detected by Northern blot hybridization with a TBSV-specific probe. −, water was added as a control; +, addition of the siRNA-containing ribonuclease fraction.
Similar articles
- Diverse and newly recognized effects associated with short interfering RNA binding site modifications on the Tomato bushy stunt virus p19 silencing suppressor.
Hsieh YC, Omarov RT, Scholthof HB. Hsieh YC, et al. J Virol. 2009 Mar;83(5):2188-200. doi: 10.1128/JVI.02186-08. Epub 2008 Dec 3. J Virol. 2009. PMID: 19052093 Free PMC article. - Biological relevance of a stable biochemical interaction between the tombusvirus-encoded P19 and short interfering RNAs.
Omarov R, Sparks K, Smith L, Zindovic J, Scholthof HB. Omarov R, et al. J Virol. 2006 Mar;80(6):3000-8. doi: 10.1128/JVI.80.6.3000-3008.2006. J Virol. 2006. PMID: 16501109 Free PMC article. - An in vitro reprogrammable antiviral RISC with size-preferential ribonuclease activity.
Omarov RT, Ciomperlik J, Scholthof HB. Omarov RT, et al. Virology. 2016 Mar;490:41-8. doi: 10.1016/j.virol.2015.12.017. Epub 2016 Jan 23. Virology. 2016. PMID: 26812224 - Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity.
Qiu W, Park JW, Scholthof HB. Qiu W, et al. Mol Plant Microbe Interact. 2002 Mar;15(3):269-80. doi: 10.1094/MPMI.2002.15.3.269. Mol Plant Microbe Interact. 2002. PMID: 11952130 - Plant RNAi: How a viral silencing suppressor inactivates siRNA.
Zamore PD. Zamore PD. Curr Biol. 2004 Mar 9;14(5):R198-200. doi: 10.1016/j.cub.2004.02.021. Curr Biol. 2004. PMID: 15028237 Review.
Cited by
- Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro.
Gago-Zachert S, Schuck J, Weinholdt C, Knoblich M, Pantaleo V, Grosse I, Gursinsky T, Behrens SE. Gago-Zachert S, et al. Nucleic Acids Res. 2019 Sep 26;47(17):9343-9357. doi: 10.1093/nar/gkz678. Nucleic Acids Res. 2019. PMID: 31433052 Free PMC article. - RNA-based viral immunity initiated by the Dicer family of host immune receptors.
Aliyari R, Ding SW. Aliyari R, et al. Immunol Rev. 2009 Jan;227(1):176-88. doi: 10.1111/j.1600-065X.2008.00722.x. Immunol Rev. 2009. PMID: 19120484 Free PMC article. Review. - An Inside Look into Biological Miniatures: Molecular Mechanisms of Viroids.
Venkataraman S, Badar U, Shoeb E, Hashim G, AbouHaidar M, Hefferon K. Venkataraman S, et al. Int J Mol Sci. 2021 Mar 10;22(6):2795. doi: 10.3390/ijms22062795. Int J Mol Sci. 2021. PMID: 33801996 Free PMC article. Review. - Deep sequencing of the small RNAs derived from two symptomatic variants of a chloroplastic viroid: implications for their genesis and for pathogenesis.
Di Serio F, Gisel A, Navarro B, Delgado S, Martínez de Alba AE, Donvito G, Flores R. Di Serio F, et al. PLoS One. 2009 Oct 21;4(10):e7539. doi: 10.1371/journal.pone.0007539. PLoS One. 2009. PMID: 19847296 Free PMC article. - Diverse and newly recognized effects associated with short interfering RNA binding site modifications on the Tomato bushy stunt virus p19 silencing suppressor.
Hsieh YC, Omarov RT, Scholthof HB. Hsieh YC, et al. J Virol. 2009 Mar;83(5):2188-200. doi: 10.1128/JVI.02186-08. Epub 2008 Dec 3. J Virol. 2009. PMID: 19052093 Free PMC article.
References
- Baulcombe D. Nature. 2004;431:356–363. - PubMed
- Scholthof HB. Nat Rev Microbiol. 2006;4:405–411. - PubMed
- Silhavy D, Burgyan J. Trends Plant Sci. 2004;9:76–83. - PubMed
- Qu F, Morris TJ. FEBS Lett. 2005;579:5958–5964. - PubMed
- Roth BM, Pruss GJ, Vance VB. Virus Res. 2004;102:97–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials