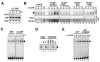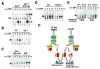A fourth IkappaB protein within the NF-kappaB signaling module - PubMed (original) (raw)
A fourth IkappaB protein within the NF-kappaB signaling module
Soumen Basak et al. Cell. 2007.
Abstract
Inflammatory NF-kappaB/RelA activation is mediated by the three canonical inhibitors, IkappaBalpha, -beta, and -epsilon. We report here the characterization of a fourth inhibitor, nfkappab2/p100, that forms two distinct inhibitory complexes with RelA, one of which mediates developmental NF-kappaB activation. Our genetic evidence confirms that p100 is required and sufficient as a fourth IkappaB protein for noncanonical NF-kappaB signaling downstream of NIK and IKK1. We develop a mathematical model of the four-IkappaB-containing NF-kappaB signaling module to account for NF-kappaB/RelA:p50 activation in response to inflammatory and developmental stimuli and find signaling crosstalk between them that determines gene-expression programs. Further combined computational and experimental studies reveal that mutant cells with altered balances between canonical and noncanonical IkappaB proteins may exhibit inappropriate inflammatory gene expression in response to developmental signals. Our results have important implications for physiological and pathological scenarios in which inflammatory and developmental signals converge.
Figures
Figure 1. LTβR induced NF-κB/RelA dimer activation does not correlate with canonical IκB kinase activity or IκB-degradation
A current model of NF-κB activation mechanisms resulting in RelA:p50 and RelB:p52 dimer activation via the canonical (IKK2) and the non-canonical (IKK1) pathway, respectively. The pathway that mediates NF-κB/RelA dimer activation in response to non-canonical stimuli has not been elucidated. B. Nuclear NF-κB DNA binding activities (arrows and arrow head) induced by 1ng/ml of TNF, 10μg/ml or 0.3μg/ml of LTβR agonistic antibody were resolved by EMSA with a κB-site containing probe. C. The composition of NF-κB DNA binding activities induced upon LTβR receptor ligation were examined by supershift analysis using the indicated antibodies (lanes 2-8). At 5hr post-stimulation, RelA:p50 (arrow) and RelB:p50 (arrowhead) dimers were detected. D. IKK kinase activity was monitored by incubating GST-IκBα with anti-NEMO co-immunoprecipitates from extracts prepared from MEF stimulated with 1ng/ml TNF, 0.01ng/ml TNF or with 10μg/ml α-LTβR antibody, 0.3μg/ml α-LTβR antibody. Reaction mixtures were resolved in SDS-PAGE and used for autoradiography (top) or immunoblotting (bottom) of co-precipitated IKK1 as a loading control. E. IκBα and -β immunoblot of extracts prepared from TNF (red lines) or LTβR (green and blue lines) stimulated cells. Signals were quantitated, normalized to the actin loading control and graphed relative to signal levels in resting cells (bottom panel).
Figure 2. nfkb2 p100 is an inhibitor of the RelA:p50 DNA binding activity
A. Subcellular localization of RelA in wild type and _i_κ_b_α−/−β−/−ε−/− MEF (_i_κ_b_−/−) was revealed by immunoblot of cytoplasmic (CE) and nuclear (NE) extracts. Signals were quantitated and respective percentages of total are indicated below the blot. An immunoblot for the TFIID component TAF20 (a.k.a. TAF12) served as a fractionation control. B. Silver-stained SDS-PAGE of tandem affinity purified (TAP-) p65 protein expressed by a retroviral transgene in _i_κ_b_α−/−β−/−ε−/−_rela_−/− MEF. Mock purification from equivalent extracts prepared from the parental MEF confirms the specificity of the associated proteins. LC-MS/MS analysis revealed that indicated gel slices contained p100, p65 and p50 proteins respectively. C. Immunoblots of RelA co-immunoprecipitates prepared from wild type (left panel) or IκB-deficient (right panel) MEF lysates revealed RelA association with IκBα, p50, p100 and p105 proteins. Lysate corresponding to half as many cells as the immunoprecipitate was probed as a control. D. The fractions of cellular p100 or p105 bound to RelA or RelB were examined by immunoblots of RelA or RelB-immunoprecipitate (IP) and compared to 50% cell-equivalent of crude lysate or flow-through (FT). E. Latent cytoplasmic NF-κB DNA binding activity in wild type (left panel) or IκB-deficient cells (right panel) were revealed by deoxycholate (DOC) treatment and EMSA (lanes 2 and 7). Prior immunodepletion with the indicated antibodies removed the target protein along with associated latent NF-κB activity. An immunoblot (bottom panel) against actin served as a loading control. F. IκB-deficient MEF were subjected to p100 “knockdown” by lentivirus mediated delivery of a specific shRNA. Lysates prepared from resulting i_κ_b_−/−_p100KD and parental _i_κ_b_−/−cells were compared by immunoblots for indicated proteins (left panel). Similarly, nuclear extracts were compared for NF-κB DNA binding activity by EMSA (right panel). G. Specificity and composition of the constitutive NF-κB DNA binding complexes present in the indicated nuclear extracts were determined by oligonucleotide competition (lanes 2-3) and supershift analysis with indicated antibodies (lanes 4-6).
Figure 3. nfkb2 p100 is a stimulus-responsive regulator of the NF-κB/RelA dimer
A. Association of p100 and RelA during LTβR signaling was monitored by p100 immunoblotting of RelA co-immunoprecipitates performed with extracts prepared from wild type cells at the indicated times. Immunoblots for p65 and p50 served as a control for co-IP efficiency. “*” indicates a non-specific band. B. The requirement of specific domains of p100 in LTβR-induced NF-κB/RelA activation was examined. NF-κB/RelA activation was scored by EMSA (top panel) and by immunoblotting (bottom panels) nuclear extracts prepared from LTβR agonist treated _nfkb2_−/− MEF or _nfkb2_−/− MEF stably transduced with retrovirus expressing full length p100, a mutant that lacks the C-terminal stimulus-responsive phosphorylation sites (“p1001-774”), a C-terminal truncated protein that lacks the IκB-like domain (“p52”), or an N-terminal truncated protein that lacks RHD (“IκBδ”). C. EMSA of NF-κB DNA binding induced in _i_κ_b_α−/−β−/−ε−/− cells by TNFR (left panel) or LTβR (right panel) stimulation for the indicated times. D. Specificity and composition of NF-κB DNA binding complexes induced by LTβR stimulation in _i_κ_b_α−/−β−/−ε−/− cells was determined by oligonucleotide competition (lanes 2–3) and supershift with the indicated antibodies (lanes 4–7). RelA- (arrow) and RelB-(arrowhead) containing complexes are indicated. E. The effect of p100 knockdown in _i_κ_b_α−/−β−/−ε−/− cells on the inducibility of NF-κB DNA binding activity by LTβR stimulation was monitored by EMSA.
Figure 4. Multiple nfkb2 p100 NF-κB complexes are functionally distinct
A. RelA interactions with p100 via the RHD or ARD were distinguished by deoxycholate (DOC) sensitivity. RelA-immunoprecipitates prepared from cytoplasmic extracts from resting (left panel) or LTβR agonist treated (right panel) cells were washed with increasing concentrations of DOC. Washes and immunoprecipitates were examined by immunoblotting with indicated antibodies. B. LTβR-induced proteolysis of pre-existing or de novo synthesized p100 protein was monitored using pulse-chase in vivo labeling, as indicated in the schematic. N-terminal p100 antibody was used in immunoprecipitates with extracts prepared from pre-labeled cells at the indicated timepoints of an LTβR timecourse (lanes 1–3, “chase”), and from cells labeled during the later phase of mock or LTβR timecourse (lanes 4–5, “pulse”). Similarly treated extracts from _nf_κ_b2_−/− cells (lane 6) served to identify non-specific bands. C. Stimulus-responsive and unresponsive p100 complexes. We propose to distinguish between self-inhibited dimeric complexes of p100 and RelA or RelB which are unresponsive to LTβR signaling, and stimulus-responsive ternary complexes in which the IκBδ domain of p100 inhibits functional RelA:p50 and RelB:p50 dimers in trans D. Increased p52 association with p100 following LTβR signaling. Immunoblot for p52/p100 proteins of p100 C-terminal domain immunoprecipitates prepared from indicated cell extracts.
Figure 5. Signal transducers of the non-canonical pathway are required for LTβR-induced RelA dimer activation
A. LTβR and TNF induced NF-κB DNA binding activities were monitored by EMSA in _ikk1_−/− MEF reconstituted with an IKK1-expressing retrovirus (lanes 5–8) and parental IKK1-deficient MEF (lanes 1–4). Arrows indicate specific NF-κB-DNA complexes. B. NF-κB DNA binding activities induced by LTβR stimulation in _nik_−/− (lanes 5–8) and control wild type (lanes 1–4) MEF was monitored by EMSA. C. NF-κB DNA binding activities induced by LTβR stimulation in _i_κ_b_α−/−β−/−ε−/− MEF (lanes 5–8) or such IκB-deficient cells in which NIK expression was “knocked down” by lentiviral shRNA expression were monitored by EMSA. D. The requirement for NEMO in LTβR activation of NF-κB was examined by EMSA of LTβR stimulated _nemo_−/− MEFs. E. Relative induction of RelA- and RelB-containing dimers was assessed by immuno-ablation of the complexes with the indicated supershift antibody. F. A schematic model of two pathways leading to the activation of NF-κB/RelA:p50 activity. The canonical IκB proteins IκBα, -β, -ε mediate RelA:p50 activation in response TNFR stimulation downstream of NEMO and IKK2. We propose that the non-canonical IκB protein p100/IκBδ inhibits the DNA binding activity of both RelA:p50 and RelB:p50 dimers and mediates their activation in response to LTβR stimulation downstream of NIK and IKK1.
Figure 6. A single NF-κB signaling module mediates NF-κB RelA activation in response to inflammatory and developmental signals
A. A schematic presentation of the NF-κB signaling module that includes three canonical IκB proteins IκBα, -β, -ε that mediate signals from IKK2 inducing stimuli, and the non-canonical IκB protein p100/δ that mediates signals from IKK1 inducing stimuli. IκBα, IκBε, and p100 are encoded by NF-κB response genes and are able to provide negative feedback. Biochemical reactions encompassed in the yellow box are represented as ordinary differential equations in a mathematical model (version 3.0, see supplementary information). B. Computational simulations of the dynamic regulation of nuclear RelA:p50 activity (top panels, red), total cellular protein levels of canonical IκB proteins (middle panels, green) and non-canonical IκB protein p100 (bottom panels, blue). Simulations are shown for the IKK2-inducing inflammatory stimuli TNF (left column) and LPS (center column) and the IKK1 inducing developmental signals mediated by LTβR (right column).
Figure 7. Nfkb2 p100 mediates crosstalk between inflammatory and developmental signals
A. Cellular levels of canonical (IκBα, -β, -ε, green) and non-canonical (nf_κ_b2 p100, blue) IκB proteins over a 20 hour timecourse as predicted by computational simulations of cells stimulated with a 1hr pulse of TNF. The pie charts indicate the proportion of NF-κB/RelA bound to canonical IκBs (green) vs. the non-canonical IκB (blue) in resting (left) vs. TNF- primed (right) cells. B. Immunoblots for canonical IκBα, and -β, and non-canonical p100/δ protein of whole cell lysates prepared at the indicated times from wild type MEF cells that were transiently stimulated with 1ng/ml of TNF for 1hr. C. EMSA for NF-κB activity liberated by deoxycholate (DOC) treatment of cytoplasmic extracts prepared from naïve (top panel) or TNF primed (bottom panel) MEF. Prior to DOC treatment extracts were either mock, IκBα, or p100/δ-immunodepleted. RelA:p50 DNA binding complexes are indicated by the arrow. D. Computational simulations of RelA:p50 activity induced by LTβR stimulation of naïve or TNF-primed cells. E. Experimental analysis of LTβR signaling in naïve or TNF primed cells. Cells were harvested at indicated time points after LTβR stimulation and nuclear extracts were tested for RelA:p50 DNA binding activity by EMSA. F. Immunoblotting of nuclear RelA protein (top) during LTβR timecourse in naïve and TNF-primed cells. An immunoblot for TAF20 (bottom) served as a loading control. G. RNAse protection assay to monitor the expression of NF-κB response genes induced by LTβR stimulation in naïve (left) and TNF primed cells (right).
Figure 8. Misregulation of canonical and non-canonical IκB homeostasis alters stimulus-specific gene expression
A. Cellular protein levels of canonical (IκBα, -β, -ε, green) and non-canonical (nf_κ_b2 p100, blue) IκB proteins in unstimulated cells of the indicated genotype as predicted by computational simulations. B. Quantitative immunoblots for p100 of lysates prepared from equivalent number of wild type or mutant MEF deficient of the indicated genotype. C. Computational simulations of nuclear RelA:p50 activity induced by LTβR stimulation in cells of the indicated genotype. D. EMSA for NF-κB binding activity of nuclear extracts prepared from LTβR stimulated cells of the indicated genotype. Wild type or mutant MEF that lack one or more IκB isoforms were stimulated with LTβR for the indicated times. E. RNAse protection assay to monitor the expression of the indicated NF-κB response genes induced by LTβR stimulation in cells of the indicated genotype.
Similar articles
- A roadmap of constitutive NF-κB activity in Hodgkin lymphoma: Dominant roles of p50 and p52 revealed by genome-wide analyses.
de Oliveira KA, Kaergel E, Heinig M, Fontaine JF, Patone G, Muro EM, Mathas S, Hummel M, Andrade-Navarro MA, Hübner N, Scheidereit C. de Oliveira KA, et al. Genome Med. 2016 Mar 17;8(1):28. doi: 10.1186/s13073-016-0280-5. Genome Med. 2016. PMID: 26988706 Free PMC article. - Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system.
Basak S, Shih VF, Hoffmann A. Basak S, et al. Mol Cell Biol. 2008 May;28(10):3139-50. doi: 10.1128/MCB.01469-07. Epub 2008 Feb 25. Mol Cell Biol. 2008. PMID: 18299388 Free PMC article. - Immune Differentiation Regulator p100 Tunes NF-κB Responses to TNF.
Chatterjee B, Roy P, Sarkar UA, Zhao M, Ratra Y, Singh A, Chawla M, De S, Gomes J, Sen R, Basak S. Chatterjee B, et al. Front Immunol. 2019 May 7;10:997. doi: 10.3389/fimmu.2019.00997. eCollection 2019. Front Immunol. 2019. PMID: 31134075 Free PMC article. - Regulation and function of IKK and IKK-related kinases.
Häcker H, Karin M. Häcker H, et al. Sci STKE. 2006 Oct 17;2006(357):re13. doi: 10.1126/stke.3572006re13. Sci STKE. 2006. PMID: 17047224 Review. - [Noncanonical NF-κB pathway and hematological malignancies].
Wang WJ, Sun AN, Guo F. Wang WJ, et al. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010 Aug;18(4):1069-73. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010. PMID: 20723331 Review. Chinese.
Cited by
- IκBβ enhances the generation of the low-affinity NFκB/RelA homodimer.
Tsui R, Kearns JD, Lynch C, Vu D, Ngo KA, Basak S, Ghosh G, Hoffmann A. Tsui R, et al. Nat Commun. 2015 May 7;6:7068. doi: 10.1038/ncomms8068. Nat Commun. 2015. PMID: 25946967 Free PMC article. - Inhibition of PHLPP2/cyclin D1 protein translation contributes to the tumor suppressive effect of NFκB2 (p100).
Xu J, Wang Y, Hua X, Xu J, Tian Z, Jin H, Li J, Wu XR, Huang C. Xu J, et al. Oncotarget. 2016 Jun 7;7(23):34112-30. doi: 10.18632/oncotarget.8746. Oncotarget. 2016. PMID: 27095572 Free PMC article. - Caspase-9b Interacts Directly with cIAP1 to Drive Agonist-Independent Activation of NF-κB and Lung Tumorigenesis.
Vu NT, Park MA, Shultz MD, Bulut GB, Ladd AC, Chalfant CE. Vu NT, et al. Cancer Res. 2016 May 15;76(10):2977-89. doi: 10.1158/0008-5472.CAN-15-2512. Epub 2016 Mar 29. Cancer Res. 2016. PMID: 27197231 Free PMC article. - Classical NF-kappaB activation negatively regulates noncanonical NF-kappaB-dependent CXCL12 expression.
Madge LA, May MJ. Madge LA, et al. J Biol Chem. 2010 Dec 3;285(49):38069-77. doi: 10.1074/jbc.M110.147207. Epub 2010 Oct 5. J Biol Chem. 2010. PMID: 20923761 Free PMC article. - Anti-Inflammatory Activity of Piquerol Isolated from Piqueria trinervia Cav.
Campos-Xolalpa N, Esquivel-Campos AL, Martínez-Casares RM, Pérez-Gutiérrez S, Pérez-Ramos J, Sánchez-Mendoza E. Campos-Xolalpa N, et al. Pharmaceuticals (Basel). 2022 Jun 21;15(7):771. doi: 10.3390/ph15070771. Pharmaceuticals (Basel). 2022. PMID: 35890070 Free PMC article.
References
- Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. - PubMed
- Browning JL, Allaire N, Ngam-Ek A, Notidis E, Hunt J, Perrin S, Fava RA. Lymphotoxin-beta receptor signaling is required for the homeostatic control of HEV differentiation and function. Immunity. 2005;23:539–550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM071573/GM/NIGMS NIH HHS/United States
- P01 GM071862/GM/NIGMS NIH HHS/United States
- R37AI33068/AI/NIAID NIH HHS/United States
- AI61549/AI/NIAID NIH HHS/United States
- CA69381/CA/NCI NIH HHS/United States
- R21 AI061549/AI/NIAID NIH HHS/United States
- GM071862/GM/NIGMS NIH HHS/United States
- R01 GM071573/GM/NIGMS NIH HHS/United States
- P01 CA069381/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous