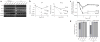DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal - PubMed (original) (raw)
DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal
Yangming Wang et al. Nat Genet. 2007 Mar.
Abstract
The molecular controls that govern the differentiation of embryonic stem (ES) cells remain poorly understood. DGCR8 is an RNA-binding protein that assists the RNase III enzyme Drosha in the processing of microRNAs (miRNAs), a subclass of small RNAs. Here we study the role of miRNAs in ES cell differentiation by generating a Dgcr8 knockout model. Analysis of mouse knockout ES cells shows that DGCR8 is essential for biogenesis of miRNAs. On the induction of differentiation, DGCR8-deficient ES cells do not fully downregulate pluripotency markers and retain the ability to produce ES cell colonies; however, they do express some markers of differentiation. This phenotype differs from that reported for Dicer1 knockout cells, suggesting that Dicer has miRNA-independent roles in ES cell function. Our findings indicate that miRNAs function in the silencing of ES cell self-renewal that normally occurs with the induction of differentiation.
Conflict of interest statement
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
Figures
Figure 1
Dgcr8 knockout strategy. The DGCR8 protein has one WW domain and two double-stranded RNA-binding domains (dsRBDs). The WW domain is encoded in exon 4 of the Dgcr8 genomic DNA (gDNA); the two dsRBDs are located in exons 7–9 and 10–12, respectively. Excision of exon 3 from the mRNA generates a frame shift, resulting in several premature stop codons in downstream exons. ES cells were sequentially targeted and treated with Cre recombinase to produce Dgcr8 Δ/flox and Δ/Δ ES cells. Rescue cells were generated by retargeting Δ/Δ ES cells with the 3lox construct, followed by removal of the HygroTK cassette with Cre recombinase.
Figure 2
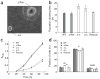
DGCR8 is essential and may be specific for miRNA biogenesis in ES cells. (a) RNA blot analysis of miRNAs from wild-type (WT), heterozygous (Δ/flox) and two independent knockout (Δ/Δ) ES cells with probes specific to miR-294 and miR-130. U6 snRNA was used as a loading control. (b) RNA blot analysis of pri-miRNA, pre-miRNA and miRNA from ES cells with a probe specific to miR-293. (c) Expression ratios of Dgcr8 heterozygous and knockout ES cells relative to wild-type ES cell for 89 miRNAs. These ratios were calculated from miRNA microarray analysis. (d) Expression ratios of Dgcr8 heterozygous and Dgcr8 knockout ES cells relative to wild-type ES cells for 19 representative miRNAs. The miRNAs on the left are ES cell–specific miRNAs; those on the right are expressed in ES cells and differentiated tissues. (e) RNA blot analysis of pre-rRNAs. Top, hybridization with a probe specific to the 5′ ETS sequence of 45S pre-rRNA (binding to 45S and 30S transcripts); bands below 30S pre-rRNA were detected owing to non-specific binding to abundant 28S rRNA. Middle, hybridization with a probe specific to the 5.8S rRNA sequence (binding to 45S, 32S, 12S and 5.8S transcripts). Bottom, U6 snRNA was used as a loading control.
Figure 3
Proliferation defects of Dgcr8 knockout ES cells. (a) Morphology of Dgcr8 Δ/flox and Δ/Δ ES cell colonies 7 d after plating. The Δ/flox colony is overgrown, with differentiation occurring around its edges. (b) Mean population doubling time (n = 5–6). (c) Mean growth rate of ES cells measured by MTT assay (n = 6). (d) Cell-cycle analysis of Dgcr8 knockout ES cells (n = 3). Error bars in b–d represent the s.d.
Figure 4
EB differentiation and teratoma formation. (a) Morphology of EBs from wild-type, Δ/Δ and rescued ES cells. EBs were cultured for 40–50 d. (b) RT-PCR analysis of pluripotency (Oct4) and differentiation markers (Fgf5,T(brachyury),Hnf4a,Krt18) after EB differentiation. Gapdh was used as a reference. (c) Quantitative RT-PCR for the markers shown in b. The β-actin gene was used as a reference. For each gene, data were normalized to the mRNA level at day 0 of wild-type EB differentiation. (d) Teratoma formation by wild-type and Dgcr8 knockout ES cells. Arrow in Δ/Δ tumor identifies region of epithelial differentiation.
Figure 5
Monolayer differentiation of ES cells in the presence of retinoic acid. (a) RT-PCR analysis of pluripotency markers. Wild-type and Dgcr8 knockout ES cells were plated as a monolayer and treated with retinoic acid (RA) in the absence of LIF to induce mesenchymal differentiation. Gapdh was used as a loading control. (b) Quantitative PCR analysis of the pluripotency markers Oct4 and Nanog (n = 3). The β-actin gene was used as a reference. For each sample, data were normalized to the mRNA level at day 0. (c) ES cell colony formation of differentiated cells. After varying durations of monolayer differentiation, cells were returned to ES cell culture conditions and assayed for their ability to form alkaline phosphatase–positive colonies. Error bars indicate the range of measurements (n = 3). (d) Clonal analysis of ES cell differentiation in the absence of MEF feeders and in the presence or absence of LIF, as indicated. Shown are percentages of undifferentiated, mixed and differentiated colonies under the indicated conditions. ‘D5’ indicates 5 d; ‘D8’ indicates 8 d.
Similar articles
- Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs.
Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Babiarz JE, et al. Genes Dev. 2008 Oct 15;22(20):2773-85. doi: 10.1101/gad.1705308. Genes Dev. 2008. PMID: 18923076 Free PMC article. - Dgcr8 knockout approaches to understand microRNA functions in vitro and in vivo.
Guo WT, Wang Y. Guo WT, et al. Cell Mol Life Sci. 2019 May;76(9):1697-1711. doi: 10.1007/s00018-019-03020-9. Epub 2019 Jan 29. Cell Mol Life Sci. 2019. PMID: 30694346 Free PMC article. Review. - Genome-wide identification of targets and function of individual MicroRNAs in mouse embryonic stem cells.
Hanina SA, Mifsud W, Down TA, Hayashi K, O'Carroll D, Lao K, Miska EA, Surani MA. Hanina SA, et al. PLoS Genet. 2010 Oct 21;6(10):e1001163. doi: 10.1371/journal.pgen.1001163. PLoS Genet. 2010. PMID: 20975942 Free PMC article. - Chromatin changes in dicer-deficient mouse embryonic stem cells in response to retinoic acid induced differentiation.
Tennakoon JB, Wang H, Coarfa C, Cooney AJ, Gunaratne PH. Tennakoon JB, et al. PLoS One. 2013 Sep 9;8(9):e74556. doi: 10.1371/journal.pone.0074556. eCollection 2013. PLoS One. 2013. PMID: 24040281 Free PMC article. - Piecing together the mosaic of early mammalian development through microRNAs.
Blakaj A, Lin H. Blakaj A, et al. J Biol Chem. 2008 Apr 11;283(15):9505-8. doi: 10.1074/jbc.R800002200. Epub 2008 Feb 13. J Biol Chem. 2008. PMID: 18272516 Free PMC article. Review.
Cited by
- Efficiency and specificity in microRNA biogenesis.
Barad O, Mann M, Chapnik E, Shenoy A, Blelloch R, Barkai N, Hornstein E. Barad O, et al. Nat Struct Mol Biol. 2012 May 13;19(6):650-2. doi: 10.1038/nsmb.2293. Nat Struct Mol Biol. 2012. PMID: 22580560 Free PMC article. - Regulatory non-coding RNAs in pluripotent stem cells.
Rosa A, Brivanlou AH. Rosa A, et al. Int J Mol Sci. 2013 Jul 11;14(7):14346-73. doi: 10.3390/ijms140714346. Int J Mol Sci. 2013. PMID: 23852015 Free PMC article. Review. - Systematically dissecting the global mechanism of miRNA functions in mouse pluripotent stem cells.
Wang A, He Q, Zhong Y. Wang A, et al. BMC Genomics. 2015 Jul 1;16(1):490. doi: 10.1186/s12864-015-1706-y. BMC Genomics. 2015. PMID: 26126859 Free PMC article. - p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells.
Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC. Jain AK, et al. PLoS Biol. 2012;10(2):e1001268. doi: 10.1371/journal.pbio.1001268. Epub 2012 Feb 28. PLoS Biol. 2012. PMID: 22389628 Free PMC article. - Tissue elasticity regulated tumor gene expression: implication for diagnostic biomarkers of primitive neuroectodermal tumor.
Vu LT, Keschrumrus V, Zhang X, Zhong JF, Su Q, Kabeer MH, Loudon WG, Li SC. Vu LT, et al. PLoS One. 2015 Mar 16;10(3):e0120336. doi: 10.1371/journal.pone.0120336. eCollection 2015. PLoS One. 2015. PMID: 25774514 Free PMC article.
References
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. - PubMed
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. - PubMed
- Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials