Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins - PubMed (original) (raw)
Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins
Eleonora Parlanti et al. Nucleic Acids Res. 2007.
Abstract
It has been hypothesized that a replication associated repair pathway operates on base damage and single strand breaks (SSB) at replication forks. In this study, we present the isolation from the nuclei of human cycling cells of a multiprotein complex containing most of the essential components of base excision repair (BER)/SSBR, including APE1, UNG2, XRCC1 and POLbeta, DNA PK, replicative POLalpha, delta and epsilon, DNA ligase 1 and cell cycle regulatory protein cyclin A. Co-immunoprecipitation revealed that in this complex DNA repair proteins are physically associated to cyclin A and to DNA replication proteins including MCM7. This complex is endowed with DNA polymerase and protein kinase activity and is able to perform BER of uracil and AP sites. This finding suggests that a preassembled DNA repair machinery is constitutively active in cycling cells and is ready to be recruited at base damage and breaks occurring at replication forks.
Figures
Figure 1.
Purification of the multiprotein DNA repair complex. (A) Elution profile of the last purification step (Mono S). POLα (black symbols), POLα/ɛ (open squares) and POLδ/ɛ (open triangles) activities coeluted in the same fractions. (B) Dot blot analysis of the Mono S fractions with specific antibodies showing coelution of cyclin A, POLα, POLδ and LIG1.
Figure 2.
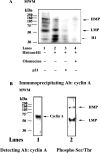
Protein kinase activity of the multiprotein complex. (A) Phosphorylated polypeptides were revealed upon incubation of the Mono S fraction in the presence of [γ-32P] ATP alone (lane 1), or in combination with histone H1 (lane 2), histone plus p21 (lane 3) or histone plus Olomucine (lane 4). (B) The Mono S fraction was immunoprecipitated with anti-cyclin A antibodies and the immunoprecipitated material was probed with antibodies against cyclin A (lane 1) or anti-phosphoSer/Thr (lane 2).
Figure 3.
Western blot analysis of the Mono S fraction. The peak fraction 14 from Mono S was analysed by immunoblotting with different antibodies against DNA repair and replication proteins, as described in material and method section. A single asterisk indicates the expected polypeptide, whereas double asterisk mark indicates degradation products.
Figure 4.
Physical association of DNA replication and repair proteins. (A) Left panel: The Mono S fraction 15 was immunoprecipitated with antibodies against cyclin A, POLβ, POLɛ or anonymous IgGs. Right panel: POLβ null (−/−) or wild type (+/+) mouse embryonic fibroblasts were lysed and the extracts used for immunoprecipitation in the presence of anti-POLβ antibodies. The immunoprecipitated material was then immunoblotted with anti-POLβ and anti-LIG1 antibodies, as indicated. S, supernatant (1:10); IP, immunoprecipitated material. (B) The Mono S fraction 15 was subjected to native gel electrophoresis, followed by immunoblotting analysis with antibodies against POLα, POLβ, POLɛ, LIG1 and XRCC1. As marked by the asterisks, all the antibodies recognized the same high molecular weight band. (C) The Mono S fraction 15 was subjected to gel filtration. Eluted proteins were analysed by dot blot with antibodies against POLα, LIG1, Cyclin A and MCM7. Arrows indicate the corresponding elution points of the molecular weight markers. (D) The gel filtration fraction 10 was analysed by western blot with antibodies against POLα, POLβ, POLδ, DNA PK, cyclin A and MCM7.
Figure 5.
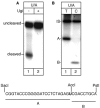
Mapping of the repair patches at AP site by the multiprotein complex. Top: autoradiograph of a denaturing polyacrylamide gel. Bottom: sequence of the restriction fragments A and B. Repair reactions were performed for 1 h in the presence of radiolabelled dTTP (lanes 1-3-5-7-9) or dCTP (lanes 2-4-6-8-10). Aphidicolin (APH), PCNA and α-POLβ were added as indicated. IS, internal standard.
Figure 6.
Incision and repair activity at uracil by the multiprotein complex. (A) Incision assay in the absence (lane 1) or presence (lane 2) of the UNG2 inhibitor Ugi. (B) Repair assay in the presence of radiolabelled dTTP (lane 1) or dCTP (lane 2). Bottom: sequence of the restriction fragments A and B. IS, internal standard.
Similar articles
- Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells.
Akbari M, Otterlei M, Peña-Diaz J, Aas PA, Kavli B, Liabakk NB, Hagen L, Imai K, Durandy A, Slupphaug G, Krokan HE. Akbari M, et al. Nucleic Acids Res. 2004 Oct 12;32(18):5486-98. doi: 10.1093/nar/gkh872. Print 2004. Nucleic Acids Res. 2004. PMID: 15479784 Free PMC article. - Post-replicative base excision repair in replication foci.
Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE. Otterlei M, et al. EMBO J. 1999 Jul 1;18(13):3834-44. doi: 10.1093/emboj/18.13.3834. EMBO J. 1999. PMID: 10393198 Free PMC article. - Monitoring of the spatial and temporal dynamics of BER/SSBR pathway proteins, including MYH, UNG2, MPG, NTH1 and NEIL1-3, during DNA replication.
Bj Rås KØ, Sousa MML, Sharma A, Fonseca DM, S Gaard CK, Bj Rås M, Otterlei M. Bj Rås KØ, et al. Nucleic Acids Res. 2017 Aug 21;45(14):8291-8301. doi: 10.1093/nar/gkx476. Nucleic Acids Res. 2017. PMID: 28575236 Free PMC article. - How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery.
Thompson LH, Hinz JM, Yamada NA, Jones NJ. Thompson LH, et al. Environ Mol Mutagen. 2005 Mar-Apr;45(2-3):128-42. doi: 10.1002/em.20109. Environ Mol Mutagen. 2005. PMID: 15668941 Review. - The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair.
Hitomi K, Iwai S, Tainer JA. Hitomi K, et al. DNA Repair (Amst). 2007 Apr 1;6(4):410-28. doi: 10.1016/j.dnarep.2006.10.004. Epub 2007 Jan 8. DNA Repair (Amst). 2007. PMID: 17208522 Review.
Cited by
- DNA-PKcs: A Targetable Protumorigenic Protein Kinase.
Dylgjeri E, Knudsen KE. Dylgjeri E, et al. Cancer Res. 2022 Feb 15;82(4):523-533. doi: 10.1158/0008-5472.CAN-21-1756. Cancer Res. 2022. PMID: 34893509 Free PMC article. Review. - Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells.
Lirussi L, Antoniali G, Vascotto C, D'Ambrosio C, Poletto M, Romanello M, Marasco D, Leone M, Quadrifoglio F, Bhakat KK, Scaloni A, Tell G. Lirussi L, et al. Mol Biol Cell. 2012 Oct;23(20):4079-96. doi: 10.1091/mbc.E12-04-0299. Epub 2012 Aug 23. Mol Biol Cell. 2012. PMID: 22918947 Free PMC article. - Physical and functional interactions between uracil-DNA glycosylase and proliferating cell nuclear antigen from the euryarchaeon Pyrococcus furiosus.
Kiyonari S, Uchimura M, Shirai T, Ishino Y. Kiyonari S, et al. J Biol Chem. 2008 Aug 29;283(35):24185-93. doi: 10.1074/jbc.M802837200. Epub 2008 Jun 18. J Biol Chem. 2008. PMID: 18562313 Free PMC article. - Glycogen Synthase Kinase 3 (GSK-3)-mediated Phosphorylation of Uracil N-Glycosylase 2 (UNG2) Facilitates the Repair of Floxuridine-induced DNA Lesions and Promotes Cell Survival.
Baehr CA, Huntoon CJ, Hoang SM, Jerde CR, Karnitz LM. Baehr CA, et al. J Biol Chem. 2016 Dec 23;291(52):26875-26885. doi: 10.1074/jbc.M116.746081. Epub 2016 Nov 14. J Biol Chem. 2016. PMID: 27875297 Free PMC article. - Mechanism and regulation of class switch recombination.
Stavnezer J, Guikema JE, Schrader CE. Stavnezer J, et al. Annu Rev Immunol. 2008;26:261-92. doi: 10.1146/annurev.immunol.26.021607.090248. Annu Rev Immunol. 2008. PMID: 18370922 Free PMC article. Review.
References
- Dogliotti E, Fortini P, Pascucci B, Parlanti E. Multiple pathways for DNA base excision repair. The mechanism of switching among multiple BER pathways. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:1–28. - PubMed
- Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J. Biol. Chem. 2001;276:5547–5555. - PubMed
- Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell. Sci. 2003;116:3051–3060. - PubMed
- Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays. 2001;23:447–455. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous