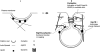A Hip1R-cortactin complex negatively regulates actin assembly associated with endocytosis - PubMed (original) (raw)
A Hip1R-cortactin complex negatively regulates actin assembly associated with endocytosis
Christophe Le Clainche et al. EMBO J. 2007.
Abstract
Actin polymerization plays a critical role in clathrin-mediated endocytosis in many cell types, but how polymerization is regulated is not known. Hip1R may negatively regulate actin assembly during endocytosis because its depletion increases actin assembly at endocytic sites. Here, we show that the C-terminal proline-rich domain of Hip1R binds to the SH3 domain of cortactin, a protein that binds to dynamin, actin filaments and the Arp2/3 complex. We demonstrate that Hip1R deleted for the cortactin-binding site loses its ability to rescue fully the formation of abnormal actin structures at endocytic sites induced by Hip1R siRNA. To determine when this complex might function during endocytosis, we performed live cell imaging. The maximum in vivo recruitment of Hip1R, clathrin and cortactin to endocytic sites was coincident, and all three proteins disappeared together upon formation of a clathrin-coated vesicle. Finally, we showed that Hip1R inhibits actin assembly by forming a complex with cortactin that blocks actin filament barbed end elongation.
Figures
Figure 1
Hip1R interacts with the SH3 domain of cortactin. (A) co-immunoprecipitation of Hip1R with cortactin from a clathrin coat extract. Cortactin immunoprecipitates (IP) and controls lacking antibody (C) were blotted with anti-Hip1R and anti-cortactin antibodies. (B) His-tagged Hip1R (40 nM) pull-down assays using GST and GST-cortactin 1–546; 1–81; 1–326; 324–546; 491–546 (10 μM). Bound (P) and unbound (S) fractions were analyzed by Western blotting (WB) using an anti-His tag antibody (upper panel) and by Coomassie blue staining (lower panel). (C) His-tagged Hip1R (1 nM) pull-down assays using the indicated concentrations of GST-cortactin. Total input (T) and bound fractions were probed with an anti-His tag antibody. The results were quantified to obtain the binding curve. The best fit of the curve gave a _K_d=20 nM. (D) Summary of the binding assay results. All the experiments were performed three times with similar results.
Figure 2
The C-terminal proline-rich region of Hip1R interacts with cortactin. (A) His-tagged Hip1R 1–1068; 346–1068 and 1–655 (400 nM) pull-down assays using GST and GST-cortactin (800 nM). Bound and unbound fractions were analyzed by Coomassie blue staining (upper panel), and by Western blotting using an anti-Hip1R antibody (lower panel). (B) His-tagged cortactin (400 nM) pull-down assays using GST and GST-Hip1R 1–312; 346–655; 766–1068 (10 μM). Bound and unbound fractions were analyzed by Coomassie blue staining (upper panel), and by Western blotting using an anti-cortactin antibody (lower panel). (C) His-tagged Hip1R 1–1017 (400 nM) pull-down assays using GST, GST-cortactin (1–546) and GST-SH3-cortactin (491–546) (800 nM). Bound and unbound fractions were analyzed by Western blotting using an anti-His tag antibody (upper panel) and by Coomassie blue staining (middle panel). The same experiment was carried out in parallel with His-tagged Hip1R 1–1068 (full length) as a positive control, bound and unbound fractions were analyzed by Western blotting using an anti-His tag antibody (lower panel). (D) Summary of the binding assay results. All experiments were performed three times with similar results.
Figure 3
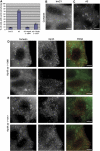
Hip1R–cortactin contributes to the regulation of actin assembly at endocytic sites. (A) The bar graph shows the percentage of HeLa cells showing abnormal actin structures after treatment with Hip1R siRNA duplex (A3), a control siRNA (InvC1), Hip1R siRNA and expression of wild-type Hip1R (A3+Hip1R 1–1068), Hip1R siRNA and expression of Hip1R 1–1017 (A3+Hip1R 1–1017). (B–E) Representative pictures of cells. (B) Cortactin staining in control siRNA (InvC1)-treated cells. (C) Cortactin staining in Hip1R siRNA (A3)-treated cells. (D–E) Representative pictures of non-rescued cells with remaining abnormal actin structures. (D) Hip1R siRNA-treated cells expressing wild-type Hip1R (1–1068). (E) Hip1R siRNA-treated cells expressing mutant Hip1R 1–1017. Cortactin staining (left panels), Hip1R staining (middle panels), merge (right panels). Scale bars are 10 μm.
Figure 4
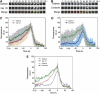
Timing of Hip1R and cortactin recruitment to CCPs. (A) Time series showing DsRed-clathrin and Hip1R-GFP recruitment to the same CCP. (B) Time series showing DsRed-clathrin and cortactin-GFP recruitment to the same CCP. (C) Average fluorescence of clathrin (dark green) and Hip1R (red) plotted against time from 30 CCPs in eight cells. The error bars represent the s.d. from 30 events. Time 0 corresponds to the moment at which the clathrin signal started to dim. All data were normalized (see Materials and methods) before averaging. (D) Average fluorescence for clathrin (light green) and cortactin (blue) plotted against time from 30 CCPs in 14 cells. (E) Summary of panels C and D without the error bars. Time 0 is marked by a yellow line.
Figure 5
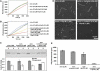
The Hip1R–cortactin complex inhibits actin assembly. (A, B) Effect of increasing amounts of Hip1R on assembly of 2.5 μM MgATP–G-actin, 10% pyrenyl-labeled, in the absence (A) or presence (B) of 0.4 μM cortactin. (C) Actin was polymerized 30 min at RT under the indicated conditions, polymerized (P) and unpolymerized (S) actin were separated by ultracentrifugation, resolved by 10% SDS–PAGE and detected by Coomassie blue (upper panel). The bar graph shows the percentage of actin in the pellets and supernatants (lower panel). The error bars represent the s.d. from three independent experiments. (D, E) Actin was polymerized in the presence of the indicated concentration of Hip1R and cortactin and the filaments were observed after 30 min by rhodamine phalloidin staining; (D) representative field for each condition; (E) quantification of the total length of polymerized actin (μm). The error bars represent the s.d. from three independent experiments.
Figure 6
The Hip1R–cortactin complex inhibits actin filament barbed end assembly. (A) Barbed end growth was measured in the presence of 100 pM spectrin–actin seeds (SP), 1 μM MgATP–G-actin, 10% pyrenyl-labeled and the indicated concentrations of Hip1R and cortactin. (B) Pointed end growth was measured in the presence of 4 nM gelsolin–actin (GA2) complex, 1.5 μM MgATP–G-actin, 10% pyrenyl-labeled and the indicated concentrations of Hip1R and cortactin. (C) The concentration of free barbed ends over time in the presence of the indicated concentrations of Hip1R and cortactin are derived from the corresponding kinetics of barbed end growth shown in panel A (Pantaloni et al, 2000) (see also Materials and methods). Inset, the relative inhibition (fraction of inhibited barbed ends corresponding to the plateau of each kinetics curve) was plotted versus the concentration of the Hip1R–cortactin complex. The binding curve for the interaction of the complex with the actin barbed end was calculated using the equation described in the Materials and methods with the best-fit value of _K_d=85 nM. (D) The fraction of barbed ends inhibited was plotted versus the total concentration of Hip1R 1–1068 (black circles), Hip1R 1–1017 (open circles) and Hip1R 346–1068 (open squares) in the presence of 0.5 μM cortactin. (E) Hip1R–cortactin inhibits actin depolymerization induced by dilution. Depolymerization of actin filaments (2 μM, 35% pyrenyl-labeled) was induced by a 40-fold dilution into polymerization buffer in the presence of indicated concentrations of Hip1R and cortactin. (F) Depolymerization rates in the presence of the indicated concentrations of Hip1R with (open squares) and without (closed circles) cortactin (25 nM). All the experiments were performed three times with the same results.
Figure 7
Hip1R inhibits Arp2/3-dependent polymerization pathways. (A) Effect of Hip1R levels on assembly of 2.5 μM MgATP–G-actin, 10% pyrenyl-labeled, in the presence of 1 μM cortactin and 350 nM Arp2/3 complex or 125 nM N-WASP and 50 nM Arp2/3 complex. (B) Effect of Hip1R levels on assembly of 2.5 μM MgATP–G-actin, 10% pyrenyl-labeled, in the presence of 50 nM cortactin, 50 nM Arp2/3 complex and 125 nM N-WASP. Inset; the maximum rate of actin assembly in the presence of Arp2/3, N-WASP and cortactin (red) or Arp2/3 and N-WASP (green) was plotted versus the concentration of Hip1R. (C, D) Reconstitution of actin assembly associated with lipid bilayers. DOPS (10 μM) liposomes containing 0.1% Texas Red DOPE were incubated with 1 μM dynamin, 1 μM cortactin, 0.5 μM Arp2/3 complex, 3 μM actin, 0.3 μM Alexa488-labeled actin and the following modifications: none (1), with 1 μM Hip1R (2), without Arp2/3 (3), without cortactin (4), without Arp2/3 and cortactin (5), without dynamin (6). Samples were observed by fluorescence microscopy. (C) Histograms showing the average fluorescence per Texas Red-labeled liposome (red) and the associated Alexa488-labeled actin cloud (green) for conditions 1–6 detailed above. The inset shows the actin/liposome ratio for the same conditions (blue). Data shown are average±s.e.m. (D) Representative images obtained for conditions (1) and (2). (a–c) Representative images obtained for liposomes (10 μM DOPS+0.1% Texas Red DOPE) incubated with 1 μM dynamin, 1 μM cortactin, 0.5 μM Arp2/3 complex, 3 μM actin, 0.3 μM Alexa488-labeled actin (condition 1). (d–f) Representative images obtained for liposomes (10 μM DOPS+0.1% Texas Red DOPE) incubated with 1 μM dynamin, 1 μM cortactin, 0.5 μM Arp2/3 complex, 3 μM actin, 0.3 μM Alexa488-labeled actin and 1 μM Hip1R (condition 2). (a, d) Liposomes; (b, e) actin; (c, f) merged. (E) His-tagged Hip1R (1 μM) pull downs using GST (1 μM) and GST-cortactin (1 μM) in the presence of dynamin (1.5 μM). Bound (P) and unbound (S) fractions were analyzed by Coomassie blue staining (upper and lower panels), bound fractions were also analyzed by Western blotting using a Hudy1 anti-dynamin antibody (middle panel). All the experiments were performed three times with the same results.
Figure 8
Model for the endocytic roles of actin, Hip1R and cortactin in endocytosis. (1) Hip1R binds to the clathrin coat, possibly stimulating or stabilizing its assembly on the inner face of the plasma membrane, and binds to actin filaments. (2) Dynamin assembles around the neck of the nascent CCV. Cortactin interacts with dynamin, stimulates the Arp2/3 complex to form branched actin filaments, and stabilizes the branched junctions. The actin meshwork grows via new filament assembly at the vesicle neck. The actin network is anchored to the clathrin coat by Hip1R, so that the vesicle is pushed away from the plasma membrane. Hip1R forms a complex with cortactin to cap actin filament barbed ends near the clathrin coat.
Similar articles
- RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery.
Engqvist-Goldstein AE, Zhang CX, Carreno S, Barroso C, Heuser JE, Drubin DG. Engqvist-Goldstein AE, et al. Mol Biol Cell. 2004 Apr;15(4):1666-79. doi: 10.1091/mbc.e03-09-0639. Epub 2004 Jan 23. Mol Biol Cell. 2004. PMID: 14742709 Free PMC article. - Regulation of cortactin/dynamin interaction by actin polymerization during the fission of clathrin-coated pits.
Zhu J, Zhou K, Hao JJ, Liu J, Smith N, Zhan X. Zhu J, et al. J Cell Sci. 2005 Feb 15;118(Pt 4):807-17. doi: 10.1242/jcs.01668. Epub 2005 Jan 25. J Cell Sci. 2005. PMID: 15671060 - The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro.
Engqvist-Goldstein AE, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG. Engqvist-Goldstein AE, et al. J Cell Biol. 2001 Sep 17;154(6):1209-23. doi: 10.1083/jcb.200106089. J Cell Biol. 2001. PMID: 11564758 Free PMC article. - Coupling actin dynamics and membrane dynamics during endocytosis.
Schafer DA. Schafer DA. Curr Opin Cell Biol. 2002 Feb;14(1):76-81. doi: 10.1016/s0955-0674(01)00297-6. Curr Opin Cell Biol. 2002. PMID: 11792548 Review. - Ever-expanding network of dynamin-interacting proteins.
Kim Y, Chang S. Kim Y, et al. Mol Neurobiol. 2006 Oct;34(2):129-36. doi: 10.1385/MN:34:2:129. Mol Neurobiol. 2006. PMID: 17220534 Review.
Cited by
- Cortactin in cancer cell migration and invasion.
Yin M, Ma W, An L. Yin M, et al. Oncotarget. 2017 Sep 19;8(50):88232-88243. doi: 10.18632/oncotarget.21088. eCollection 2017 Oct 20. Oncotarget. 2017. PMID: 29152154 Free PMC article. Review. - Erythroblast enucleation.
Keerthivasan G, Wickrema A, Crispino JD. Keerthivasan G, et al. Stem Cells Int. 2011;2011:139851. doi: 10.4061/2011/139851. Epub 2011 Oct 5. Stem Cells Int. 2011. PMID: 22007239 Free PMC article. - Endocytosis and signaling: cell logistics shape the eukaryotic cell plan.
Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, Di Fiore PP. Sigismund S, et al. Physiol Rev. 2012 Jan;92(1):273-366. doi: 10.1152/physrev.00005.2011. Physiol Rev. 2012. PMID: 22298658 Free PMC article. Review. - Load adaptation by endocytic actin networks.
Kaplan C, Kenny SJ, Chen X, Schöneberg J, Sitarska E, Diz-Muñoz A, Akamatsu M, Xu K, Drubin DG. Kaplan C, et al. Mol Biol Cell. 2022 May 15;33(6):ar50. doi: 10.1091/mbc.E21-11-0589. Epub 2022 Apr 7. Mol Biol Cell. 2022. PMID: 35389747 Free PMC article. - Vesicle trafficking plays a novel role in erythroblast enucleation.
Keerthivasan G, Small S, Liu H, Wickrema A, Crispino JD. Keerthivasan G, et al. Blood. 2010 Oct 28;116(17):3331-40. doi: 10.1182/blood-2010-03-277426. Epub 2010 Jul 19. Blood. 2010. PMID: 20644112 Free PMC article.
References
- Barroso C, Rodenbusch SE, Welch MD, Drubin DG (2006) A role for cortactin in Listeria monocytogenes invasion of NIH 3T3 cells, but not in its intracellular motility. Cell Motil Cytoskeleton 63: 231–243 - PubMed
- Benesch S, Polo S, Lai FP, Anderson KI, Stradal TE, Wehland J, Rottner K (2005) N-WASP deficiency impairs EGF internalization and actin assembly at clathrin-coated pits. J Cell Sci 118: 3103–3115 - PubMed
- Cao H, Weller S, Orth JD, Chen J, Huang B, Chen JL, Stamnes M, McNiven MA (2005) Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat Cell Biol 7: 483–492 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous