Ink4a/Arf tumor suppressor does not modulate the degenerative conditions or tumor spectrum of the telomerase-deficient mouse - PubMed (original) (raw)
Ink4a/Arf tumor suppressor does not modulate the degenerative conditions or tumor spectrum of the telomerase-deficient mouse
Christine M Khoo et al. Proc Natl Acad Sci U S A. 2007.
Abstract
The Rb/p16(Ink4a) and p53/p19Arf tumor suppressor pathways have been linked to diverse cancer-relevant processes, including those governing the cellular responses to telomere dysfunction. In this study, we sought to provide direct genetic evidence of a role for the Ink4a/Arf tumor suppressor gene, encoding both p16(Ink4a) and p19(Arf), in modulating the cellular and tissue phenotypes associated with telomere dysfunction by using the mTerc Ink4a/Arf mouse model. In contrast to the rescue associated with p53 deficiency, Ink4a/Arf deficiency did not attenuate the degenerative phenotypes elicited by telomere dysfunction in the late-generation mTerc-/- mice. Furthermore, in contrast to accelerated cancer onset and increased epithelial cancers of late-generation mTerc-/- p53 mutant mice, late-generation mTerc-/- Ink4a/Arf mutant mice experienced a delayed tumor onset and maintained the lymphoma and sarcoma spectrum. Consistent with the negligible role of Ink4a/Arf in the telomere checkpoint response in vivo, late-generation mTerc-/- Ink4a/Arf-/- tissues show activated p53, and derivative tumor cell lines sustain frequent loss of p53 function, whereas all early generation mTerc Ink4a/Arf-/- tumor cell lines remain intact for p53. In addition, the late-generation mTerc-/- Ink4a/Arf-/- tumors showed activation of the alternative lengthening of telomere mechanism, underscoring the need for adaptation to the presence of telomere dysfunction in the absence of p16(Ink4a) and p19(Arf). These observations highlight the importance of genetic context in dictating whether telomere dysfunction promotes or suppresses age-related degenerative conditions as well as the rate of initiation and type of spontaneous cancers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
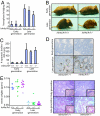
Loss of p16Ink4a and p19Arf does not modulate the in vivo phenotype of telomere dysfunction. (A) Increase in the percentage of anaphases with anaphase bridging in the GI tract of late-generation _mTerc_−/− mice. Error bars represent mean ± SE (n = 4–7 for each group). No significant difference between late-generation _mTerc_−/− Ink4a/Arf+/+ and −/− mice (P = 0.1306). Twenty-five anaphases were counted per sample. (B) Representative examples of the degenerative phenotype seen in 12- to 13-week-old G4 _mTerc_−/− Ink4a/Arf mutant mice compared with age-matched G0 mTerc+/− Ink4a/Arf controls. (C) Increase in apoptotic bodies in the crypts of the GI tract in late-generation mTerc mutant mice with different Ink4a/Arf genotypes. No significant differences between the number of apoptotic bodies between the different Ink4a/Arf genotypes within the same generations (P = 0.9084). Error bars represent mean ± SE (n = 4–7 for each group). (D) p53 induction in late-generation _mTerc_−/− Ink4a/Arf+/+ and Ink4a/_Arf_−/− intestinal crypts. (E) Testes weights (Left) and histological sections (Right) of early and late-generation mTerc mutant mice with different Ink4a/Arf genotypes. Loss of Ink4a/Arf does not rescue germ cell depletion in late-generation _mTerc_−/− testes. Inset shows depletion of spermatogonia in late-generation _mTerc_−/− testes.
Fig. 2.
Telomere dysfunction suppresses the incidence and delays the onset of tumors from the mTerc Ink4a/Arf mutant mice. (A) Kaplan–Meier analysis of tumor incidence of early generation mTerc mice with differing Ink4a/Arf genotypes (n = 34 for Ink4a/Arf+/+; n = 50 for Ink4a/Arf+/−; n = 62 for Ink4a/_Arf_−/−). (B) Kaplan–Meier analysis showing an increase of tumor latency in late-generation mTerc Ink4a/Arf+/− mice (n = 50 for early generation; n = 72 for late generation). (C) Kaplan–Meier analysis showing an increase of tumor latency in late-generation mTerc Ink4a/_Arf_−/− mice (n = 62 for early generation; n = 64 for late generation). Mice with multiple tumors were counted once.
Fig. 3.
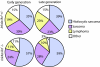
Telomere dysfunction did not change the tumor spectrums of the mTerc Ink4a/Arf mutant mice. Tumors were diagnosed histologically and graphed by percentages. Multiple tumors from the same mouse were counted as separate incidences (n = 38 for early generation mTerc Ink4a/_Arf_−/− mice; n = 27 for early generation mTerc Ink4a/Arf+/− mice; n = 13 for late-generation _mTerc_−/− Ink4a/Arf+/− mice; n = 29 for late-generation _mTerc_−/− Ink4a/_Arf_−/− mice).
Fig. 4.
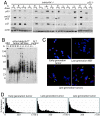
p53 checkpoint function and activation of ALT in the soft tissue sarcomas. (A) Compromised induction of p53 and p21, and Ser-18 phosphorylation of p53 in response to doxorubicin in a subset of the soft tissue sarcoma cell lines from late-generation _mTerc_−/− Ink4a/_Arf_−/− mice (L) compared with those from early generation mTerc Ink4a/_Arf_−/− mice (E). Cell lines were treated with 0.5 μg/ml doxorubicin and harvested at 0 and 16 h. Actin was used as loading control. (B) Telomere repeat fragment Southern of mTerc Ink4a/_Arf_−/− tumor cell lines showing activation of ALT in late-generation _mTerc_−/− Ink4a/_Arf_−/− tumors (L). Note increase in telomere length heterogeneity in late-generation _mTerc_−/− Ink4a/_Arf_−/− tumor cell lines (L). (C) Representative metaphase spreads of one early generation and two different late-generation sarcoma cell lines showing chromosomal fusions and heterogeneous telomere length in late-generation tumor cell lines. Late-generation _mTerc_−/− Ink4a/_Arf_−/− MEF with homogeneous short telomeres shown for comparison. Chromosomes were stained with DAPI (blue), and telomeric DNA was detected by FISH with Cy3-conjugated T2AG3 peptide nucleic acid probe (red). (D) Telomere length analysis by quantitative telomeric FISH of an early generation sarcoma cell line and two different late-generation sarcoma cell lines. Percent telomeres were plotted against TFU. Mean TFU and percent signal-free ends were 103.46, 181.94, and 141.96, and 0.9, 9.3, and 8.7%, respectively.
Fig. 5.
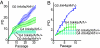
Telomere dysfunction partially suppresses the growth and immortalization of Ink4a/Arf_−/− MEFs and astrocytes. (A) 3T3 growth assay of G0 and G4 m_Terc Ink4a/Arf MEFs. (B) 3T3 growth assay of G0 and G4 mTerc Ink4a/Arf mouse embryonic astrocytes (n = 2 each for G0 mTerc+/− Ink4a/Arf+/+ and Ink4a/_Arf_−/−; n = 3 each for G4 _mTerc_−/− Ink4a/Arf+/+ and Ink4a/_Arf_−/− for both MEFs and astrocytes).
Similar articles
- Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression.
Chang S, Khoo CM, Naylor ML, Maser RS, DePinho RA. Chang S, et al. Genes Dev. 2003 Jan 1;17(1):88-100. doi: 10.1101/gad.1029903. Genes Dev. 2003. PMID: 12514102 Free PMC article. - The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis.
Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. Sharpless NE, et al. Oncogene. 2004 Jan 15;23(2):379-85. doi: 10.1038/sj.onc.1207074. Oncogene. 2004. PMID: 14724566 - Tumor escape in a Wnt1-dependent mouse breast cancer model is enabled by p19Arf/p53 pathway lesions but not p16 Ink4a loss.
Debies MT, Gestl SA, Mathers JL, Mikse OR, Leonard TL, Moody SE, Chodosh LA, Cardiff RD, Gunther EJ. Debies MT, et al. J Clin Invest. 2008 Jan;118(1):51-63. doi: 10.1172/JCI33320. J Clin Invest. 2008. PMID: 18060046 Free PMC article. - Genetic dissection of melanoma pathways in the mouse.
Yang FC, Merlino G, Chin L. Yang FC, et al. Semin Cancer Biol. 2001 Jun;11(3):261-8. doi: 10.1006/scbi.2000.0376. Semin Cancer Biol. 2001. PMID: 11407950 Review. - p53-Dependent and -independent functions of the Arf tumor suppressor.
Sherr CJ, Bertwistle D, DEN Besten W, Kuo ML, Sugimoto M, Tago K, Williams RT, Zindy F, Roussel MF. Sherr CJ, et al. Cold Spring Harb Symp Quant Biol. 2005;70:129-37. doi: 10.1101/sqb.2005.70.004. Cold Spring Harb Symp Quant Biol. 2005. PMID: 16869746 Review.
Cited by
- Telomere shortening accelerates tumor initiation in the L2-IL1B mouse model of Barrett esophagus and emerges as a possible biomarker.
Sahm V, Maurer C, Baumeister T, Anand A, Strangmann J, Schmid RM, Wang TC, Quante M. Sahm V, et al. Oncotarget. 2022 Feb 14;13:347-359. doi: 10.18632/oncotarget.28198. eCollection 2022. Oncotarget. 2022. PMID: 35178191 Free PMC article. - Telomere dysfunction alters intestinal stem cell dynamics to promote cancer.
LaBella KA, Hsu WH, Li J, Qi Y, Liu Y, Liu J, Wu CC, Liu Y, Song Z, Lin Y, Blecher JM, Jiang S, Shang X, Han J, Spring DJ, Zhang J, Xia Y, DePinho RA. LaBella KA, et al. Dev Cell. 2024 Jun 3;59(11):1475-1486.e5. doi: 10.1016/j.devcel.2024.03.020. Epub 2024 Apr 3. Dev Cell. 2024. PMID: 38574731 - Control of Cellular Aging, Tissue Function, and Cancer by p53 Downstream of Telomeres.
Roake CM, Artandi SE. Roake CM, et al. Cold Spring Harb Perspect Med. 2017 May 1;7(5):a026088. doi: 10.1101/cshperspect.a026088. Cold Spring Harb Perspect Med. 2017. PMID: 28289249 Free PMC article. Review. - Telomere dysfunction and tumour suppression: the senescence connection.
Deng Y, Chan SS, Chang S. Deng Y, et al. Nat Rev Cancer. 2008 Jun;8(6):450-8. doi: 10.1038/nrc2393. Nat Rev Cancer. 2008. PMID: 18500246 Free PMC article. Review. - Power of PTEN/AKT: Molecular switch between tumor suppressors and oncogenes.
Xie Y, Naizabekov S, Chen Z, Tokay T. Xie Y, et al. Oncol Lett. 2016 Jul;12(1):375-378. doi: 10.3892/ol.2016.4636. Epub 2016 May 26. Oncol Lett. 2016. PMID: 27347153 Free PMC article.
References
- Shay JW, Pereira-Smith OM, Wright WE. Exp Cell Res. 1991;196:33–39. - PubMed
- Hara E, Tsurui H, Shinozaki A, Nakada S, Oda K. Biochem Biophys Res Commun. 1991;179:528–534. - PubMed
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. Science. 1999;283:1321–1325. - PubMed
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. Cell. 1999;97:527–538. - PubMed
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Nature. 2000;406:641–645. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous