A conditional yeast E1 mutant blocks the ubiquitin-proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome - PubMed (original) (raw)
A conditional yeast E1 mutant blocks the ubiquitin-proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome
Nazli Ghaboosi et al. Mol Biol Cell. 2007 May.
Abstract
E1 ubiquitin activating enzyme catalyzes the initial step in all ubiquitin-dependent processes. We report the isolation of uba1-204, a temperature-sensitive allele of the essential Saccharomyces cerevisiae E1 gene, UBA1. Uba1-204 cells exhibit dramatic inhibition of the ubiquitin-proteasome system, resulting in rapid depletion of cellular ubiquitin conjugates and stabilization of multiple substrates. We have employed the tight phenotype of this mutant to investigate the role ubiquitin conjugates play in the dynamic interaction of the UbL/UBA adaptor proteins Rad23 and Dsk2 with the proteasome. Although proteasomes purified from mutant cells are intact and proteolytically active, they are depleted of ubiquitin conjugates, Rad23, and Dsk2. Binding of Rad23 to these proteasomes in vitro is enhanced by addition of either free or substrate-linked ubiquitin chains. Moreover, association of Rad23 with proteasomes in mutant and wild-type cells is improved upon stabilizing ubiquitin conjugates with proteasome inhibitor. We propose that recognition of polyubiquitin chains by Rad23 promotes its shuttling to the proteasome in vivo.
Figures
Figure 1.
Uba1-204 cells are temperature-sensitive and undergo cell cycle arrest. Ten-fold serial dilutions of wild-type and uba1-204 yeast cells were spotted onto YPD (A) and SD plates containing 30 μM CdCl2 (B) and incubated at the indicated temperatures for 2–3 d. (C) Wild-type and uba1-204 cells were arrested with α-factor for 2 h at the permissive temperature. The temperature was shifted to 37°C for 1 h, and the cells were released into fresh medium at 37°C at time 0. At indicated time points after release, aliquots were withdrawn and prepared for assessment of total DNA content by fluorescence-activated cell sorting. (D) WT and uba1-204 cells were arrested with α-factor for 3 h at the permissive temperature. Cells were then released into fresh medium and grown at 25°C for 90 min to allow progression to G2 phase. Cells were then shifted to 37°C at time 0, and aliquots were withdrawn and analyzed as described in C.
Figure 2.
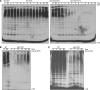
Uba1-204 cells are defective in ubiquitin conjugation. (A) Wild-type and uba1-204 cells were grown to log phase in liquid YPD medium at 25°C. Half of each culture was shifted to the nonpermissive temperature at time 0, and samples were withdrawn every minute. Lysates were prepared and analyzed by SDS-PAGE and immunoblotted with antiserum to ubiquitin. (B) Loss of ubiquitin conjugation is reversible. Strains were grown as described in A, and samples were shifted to the nonpermissive temperature for 1 h. At time 0, the temperature was shifted back to 25°C, aliquots were withdrawn at indicated time points, and ubiquitin conjugation was assessed as described in A. (C) Uba1-204 cells are active in ubiquitin conjugation at 4°C. Cells were grown in YPD medium and shifted to 37°C for 1 h. At time 0, ice was added to cultures, and the temperature was shifted to 4°C. Samples were withdrawn and analyzed as described in A.
Figure 3.
Uba1-204 cells are defective in substrate degradation. (A) Wild-type and uba1-204 cells expressing Deg1-GFP were grown in SD medium. Cultures of exponentially growing cells were incubated at 25°C or shifted to 37°C for 1 h, and cycloheximide was added to initiate a chase period. Samples were withdrawn every 15 min for analysis. Lysates were separated by SDS-PAGE and immunoblotted with antiserum to GFP. Control (−) sample was an isogenic strain lacking the plasmid. (B) WT and uba1-204 cells expressing UbV76-V-βgal under control of the GAL1 promoter were grown in SD medium with raffinose, and protein expression was induced by addition of 2% galactose for 1 h. Cultures were then incubated at either 25°C or shifted to 37°C for 1 h and transferred to dextrose medium pre-equilibrated at the same temperature to initiate a chase. Samples were withdrawn every 15 min, lysed, separated by SDS-PAGE, and immunoblotted with antiserum to β-gal. (C) WT and uba1-204 cells expressing Sic1 under the control of the GAL1 promoter were grown in SD medium with raffinose, and they were arrested with α-factor for 2 h. Sic1 expression was induced by addition of galactose to 2%, and cells were incubated at 25°C for 1 h before shifting to 37°C for an additional hour. Cells were then transferred to dextrose medium to initiate a chase, and samples were withdrawn every 30 min, lysed, separated by SDS-PAGE, and immunoblotted with antiserum to Sic1. Protein loading was verified by immunoblotting with antiserum to Cdc28.
Figure 4.
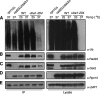
26S proteasomes isolated from uba1-204 cells are properly assembled and proteolytically active. (A) Intact 26S proteasomes can be purified from uba1-204 cells. Wild-type and uba1-204 cells were incubated at 25 or 37°C for 40 min, and 26S proteasomes were affinity purified as described in Materials and Methods. Samples were resolved by SDS-PAGE and visualized by Coomassie blue staining. 19S and 20S subunits are specified. (B) Purified proteasome complexes were resolved by nondenaturing PAGE and were visualized by Coomassie blue staining. CP refers to 20S core particle, and R1P and R2P refer to 26S proteasomes with one or two regulatory caps, respectively. (C) Proteolytic activity of the complexes in the nondenaturing gel from B was detected by fluorogenic peptide overlay with the peptidase substrate Suc-LLVY-AMC. Fluorescent bands were visualized by exposure to UV light. (D) Uba1-204 26S proteasomes are active in the degradation and deubiquitination of Ub-Sic1 in vitro. 26S proteasomes purified from wild-type and uba1-204 cells were incubated with Ub-Sic1 at 30°C for 5 min. For deubiquitination reactions, proteasomes were preincubated with 100 μM epoxomicin for 30 min at 30°C before incubation with Ub-Sic1. Samples were analyzed by SDS-PAGE followed by immunoblotting with anti-Sic1 antibody. Polyubiquitinated Sic1 (Ub-Sic1) and deubiquitinated Sic1 are specified. There is a band of unknown identity (*) that was reproducibly detected in the mutant but not wild-type lanes in the presence of epoxomicin.
Figure 5.
26S proteasomes isolated from uba1-204 cells exhibit reduced content of UbL/UBA proteins. (A–E) Wild-type and uba1-204 cells expressing Pre1-FH were grown in casamino acid medium, and half the cultures were shifted to 37°C for 40 min. Lysates were prepared and immunoprecipitated as described in Materials and Methods by incubation with anti-Flag resin in the presence of ATP. Intact 26S proteasomes were eluted with Flag peptide, separated by SDS-PAGE, and proteasome-bound proteins were detected by immunoblotting with antiserum to the specified proteins. Antibody specificity was verified using deletion mutants.
Figure 6.
Polyubiquitin chains promote Rad23 binding to the proteasome. (A) Ub-Sic1 promotes association of full-length GST-Rad23 with proteasome. Sepharose-immobilized proteasomes isolated from uba1-204 cells were preincubated with proteasome inhibitors for 1 h and then incubated with GST fusion proteins in the presence or absence of Ub-Sic1 as described in Materials and Methods. GST-Rad23 refers to the full-length protein, whereas GST-UBA contains both UBA domains and lacks the UbL domain. GST-UbL contains only the N-terminal UbL domain and was detected with antiserum to GST. The presence of proteasome complexes was verified with antibodies to Rpn10. As a negative control, simultaneous purification was conducted with uba1-204 cells lacking the Pre1-Flag allele (UT). (B) Ub-Sic1 promotes association of Rad23-HIS with proteasome. Immobilized proteasomes were prepared as described in A and incubated with Rad23-HIS in the presence or absence of Ub-Sic1. (C) Free tetraubiquitin chains are sufficient to promote GST–Rad23 association. Immobilized proteasomes were prepared as described in A and incubated with GST-Rad23 and Ub4 rather than Ub-Sic1. (D) Stabilization of polyubiquitin conjugates in vivo promotes Rad23 targeting to the proteasome. Wild-type and uba1-204 cells were treated with MG132 or DMSO (−) and shifted to 37°C for 1 h. Proteasomes were isolated by affinity purification, ubiquitin conjugates were detected with antiserum to ubiquitin, and Rad23 binding was assessed.
Similar articles
- Yeast Pth2 is a UBL domain-binding protein that participates in the ubiquitin-proteasome pathway.
Ishii T, Funakoshi M, Kobayashi H. Ishii T, et al. EMBO J. 2006 Nov 29;25(23):5492-503. doi: 10.1038/sj.emboj.7601418. Epub 2006 Nov 2. EMBO J. 2006. PMID: 17082762 Free PMC article. - In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome.
Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, Saeki Y. Tsuchiya H, et al. Mol Cell. 2017 May 18;66(4):488-502.e7. doi: 10.1016/j.molcel.2017.04.024. Mol Cell. 2017. PMID: 28525741 - Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Chen X, Randles L, Shi K, Tarasov SG, Aihara H, Walters KJ. Chen X, et al. Structure. 2016 Aug 2;24(8):1257-1270. doi: 10.1016/j.str.2016.05.018. Epub 2016 Jul 7. Structure. 2016. PMID: 27396824 Free PMC article. - Rad23 and Rpn10: perennial wallflowers join the melee.
Madura K. Madura K. Trends Biochem Sci. 2004 Dec;29(12):637-40. doi: 10.1016/j.tibs.2004.10.008. Trends Biochem Sci. 2004. PMID: 15544949 Review. - The moonlighting of RAD23 in DNA repair and protein degradation.
Grønbæk-Thygesen M, Kampmeyer C, Hofmann K, Hartmann-Petersen R. Grønbæk-Thygesen M, et al. Biochim Biophys Acta Gene Regul Mech. 2023 Jun;1866(2):194925. doi: 10.1016/j.bbagrm.2023.194925. Epub 2023 Feb 28. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 36863450 Review.
Cited by
- The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously.
Lee TV, Ding T, Chen Z, Rajendran V, Scherr H, Lackey M, Bolduc C, Bergmann A. Lee TV, et al. Development. 2008 Jan;135(1):43-52. doi: 10.1242/dev.011288. Epub 2007 Nov 28. Development. 2008. PMID: 18045837 Free PMC article. - Yeast Irc22 Is a Novel Dsk2-Interacting Protein that Is Involved in Salt Tolerance.
Ishii T, Funakoshi M, Kobayashi H, Sekiguchi T. Ishii T, et al. Cells. 2014 Mar 27;3(2):180-98. doi: 10.3390/cells3020180. Cells. 2014. PMID: 24709957 Free PMC article. - Degradation Signals for Ubiquitin-Proteasome Dependent Cytosolic Protein Quality Control (CytoQC) in Yeast.
Maurer MJ, Spear ED, Yu AT, Lee EJ, Shahzad S, Michaelis S. Maurer MJ, et al. G3 (Bethesda). 2016 Jul 7;6(7):1853-66. doi: 10.1534/g3.116.027953. G3 (Bethesda). 2016. PMID: 27172186 Free PMC article. - Components of the ubiquitin-proteasome pathway compete for surfaces on Rad23 family proteins.
Goh AM, Walters KJ, Elsasser S, Verma R, Deshaies RJ, Finley D, Howley PM. Goh AM, et al. BMC Biochem. 2008 Jan 30;9:4. doi: 10.1186/1471-2091-9-4. BMC Biochem. 2008. PMID: 18234089 Free PMC article. - Functional rescue of mutant human cystathionine beta-synthase by manipulation of Hsp26 and Hsp70 levels in Saccharomyces cerevisiae.
Singh LR, Kruger WD. Singh LR, et al. J Biol Chem. 2009 Feb 13;284(7):4238-45. doi: 10.1074/jbc.M806387200. Epub 2008 Dec 12. J Biol Chem. 2009. PMID: 19074437 Free PMC article.
References
- Amerik A. Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochem. Biophys. Acta. 2004;1695:189–207. - PubMed
- Babbitt S. E., et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. - PubMed
- Bertolaet B. L., Clarke D. J., Wolff M., Watson M. H., Henze M., Divita G., Reed S. I. UBA domains mediate protein-protein interactions between two DNA damage–inducible proteins. J. Mol. Biol. 2001a;313:955–963. - PubMed
- Bertolaet B. L., Clarke D. J., Wolff M., Watson M. H., Henze M., Divita G., Reed S. I. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Mol. Biol. 2001b;8:417–422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous