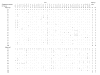Sequence variation in human succinate dehydrogenase genes: evidence for long-term balancing selection on SDHA - PubMed (original) (raw)
Sequence variation in human succinate dehydrogenase genes: evidence for long-term balancing selection on SDHA
Bora E Baysal et al. BMC Biol. 2007.
Abstract
Background: Balancing selection operating for long evolutionary periods at a locus is characterized by the maintenance of distinct alleles because of a heterozygote or rare-allele advantage. The loci under balancing selection are distinguished by their unusually high polymorphism levels. In this report, we provide statistical and comparative genetic evidence suggesting that the SDHA gene is under long-term balancing selection. SDHA encodes the major catalytical subunit (flavoprotein, Fp) of the succinate dehydrogenase enzyme complex (SDH; mitochondrial complex II). The inhibition of Fp by homozygous SDHA mutations or by 3-nitropropionic acid poisoning causes central nervous system pathologies. In contrast, heterozygous mutations in SDHB, SDHC, and SDHD, the other SDH subunit genes, cause hereditary paraganglioma (PGL) tumors, which show constitutive activation of pathways induced by oxygen deprivation (hypoxia).
Results: We sequenced the four SDH subunit genes (10.8 kb) in 24 African American and 24 European American samples. We also sequenced the SDHA gene (2.8 kb) in 18 chimpanzees. Increased nucleotide diversity distinguished the human SDHA gene from its chimpanzee ortholog and from the PGL genes. Sequence analysis uncovered two common SDHA missense variants and refuted the previous suggestions that these variants originate from different genetic loci. Two highly dissimilar SDHA haplotype clusters were present in intermediate frequencies in both racial groups. The SDHA variation pattern showed statistically significant deviations from neutrality by the Tajima, Fu and Li, Hudson-Kreitman-Aguadé, and Depaulis haplotype number tests. Empirically, the elevated values of the nucleotide diversity (% pi = 0.231) and the Tajima statistics (D = 1.954) in the SDHA gene were comparable with the most outstanding cases for balancing selection in the African American population.
Conclusion: The SDHA gene has a strong signature of balancing selection. The SDHA variants that have increased in frequency during human evolution might, by influencing the regulation of cellular oxygen homeostasis, confer protection against certain environmental toxins or pathogens that are prevalent in Africa.
Figures
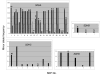
Figure 1
Minor allele frequency of each SDH subunit variant (also see Additional Table 1) is shown. Filled vertical bars refer to African American, unfilled vertical bars refer to European American samples. Synonymous and non-synonymous coding variants are marked by S and NS, respectively. ID refers to insertion/deletion polymorphisms.
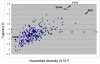
Figure 2
Tajima's D statistic and nucleotide diversity in SDHA and SDHB genes (squares) relative to the 282 genes in SeattleSNP database are shown in the African American population. Positions of two highly polymorphic loci, ABO and FUT2, the latter of which encodes the alpha(1,2)fucosyltransferase, are shown.
Figure 3
Haplotypes of SDHA. Periods denote the identical SNP variant when compared to the most common haplotype of each haplogroup. @NC = non-coding, S = synonymous coding, NS = non-synonymous coding. I = insertion allele, D = deletion allele. ^AA = African-American samples, EA = European-American samples.
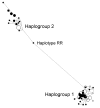
Figure 4
A median-joining network groups all SDHA haplotypes (Figure 3) on the basis of number of nucleotide differences. The haplotype RR is probably a recombinant between the two haplogroups. The pie chart for each haplotype depicts the proportional contribution of the African American (filled portion) and the European American (unfilled portion) samples.
Similar articles
- SDHA is a tumor suppressor gene causing paraganglioma.
Burnichon N, Brière JJ, Libé R, Vescovo L, Rivière J, Tissier F, Jouanno E, Jeunemaitre X, Bénit P, Tzagoloff A, Rustin P, Bertherat J, Favier J, Gimenez-Roqueplo AP. Burnichon N, et al. Hum Mol Genet. 2010 Aug 1;19(15):3011-20. doi: 10.1093/hmg/ddq206. Epub 2010 May 18. Hum Mol Genet. 2010. PMID: 20484225 Free PMC article. - Analysis of all subunits, SDHA, SDHB, SDHC, SDHD, of the succinate dehydrogenase complex in KIT/PDGFRA wild-type GIST.
Pantaleo MA, Astolfi A, Urbini M, Nannini M, Paterini P, Indio V, Saponara M, Formica S, Ceccarelli C, Casadio R, Rossi G, Bertolini F, Santini D, Pirini MG, Fiorentino M, Basso U, Biasco G; GIST Study Group. Pantaleo MA, et al. Eur J Hum Genet. 2014 Jan;22(1):32-9. doi: 10.1038/ejhg.2013.80. Epub 2013 Apr 24. Eur J Hum Genet. 2014. PMID: 23612575 Free PMC article. - Mitochondrial complex II and genomic imprinting in inheritance of paraganglioma tumors.
Baysal BE. Baysal BE. Biochim Biophys Acta. 2013 May;1827(5):573-7. doi: 10.1016/j.bbabio.2012.12.005. Epub 2013 Jan 2. Biochim Biophys Acta. 2013. PMID: 23291190 Review. - SDHA Germline Mutations in SDH-Deficient GISTs: A Current Update.
Schipani A, Nannini M, Astolfi A, Pantaleo MA. Schipani A, et al. Genes (Basel). 2023 Mar 4;14(3):646. doi: 10.3390/genes14030646. Genes (Basel). 2023. PMID: 36980917 Free PMC article. Review.
Cited by
- SDHA is a tumor suppressor gene causing paraganglioma.
Burnichon N, Brière JJ, Libé R, Vescovo L, Rivière J, Tissier F, Jouanno E, Jeunemaitre X, Bénit P, Tzagoloff A, Rustin P, Bertherat J, Favier J, Gimenez-Roqueplo AP. Burnichon N, et al. Hum Mol Genet. 2010 Aug 1;19(15):3011-20. doi: 10.1093/hmg/ddq206. Epub 2010 May 18. Hum Mol Genet. 2010. PMID: 20484225 Free PMC article. - Sdha+/- Rats Display Minimal Muscle Pathology Without Significant Behavioral or Biochemical Abnormalities.
Siebers EM, Choi MJ, Tinklenberg JA, Beatka MJ, Ayres S, Meng H, Helbling DC, Takizawa A, Bennett B, Garces AM, Dias Duarte Machado LG, Dimmock D, Dwinell MR, Geurts AM, Lawlor MW. Siebers EM, et al. J Neuropathol Exp Neurol. 2018 Aug 1;77(8):665-672. doi: 10.1093/jnen/nly042. J Neuropathol Exp Neurol. 2018. PMID: 29850869 Free PMC article. - Evolutionary and functional properties of a two-locus beta-globin polymorphism in Indian house mice.
Runck AM, Weber RE, Fago A, Storz JF. Runck AM, et al. Genetics. 2010 Apr;184(4):1121-31. doi: 10.1534/genetics.109.113506. Epub 2010 Jan 25. Genetics. 2010. PMID: 20100937 Free PMC article. - Biochemical, Molecular, and Clinical Characterization of Succinate Dehydrogenase Subunit A Variants of Unknown Significance.
Bannon AE, Kent J, Forquer I, Town A, Klug LR, McCann K, Beadling C, Harismendy O, Sicklick JK, Corless C, Shinde U, Heinrich MC. Bannon AE, et al. Clin Cancer Res. 2017 Nov 1;23(21):6733-6743. doi: 10.1158/1078-0432.CCR-17-1397. Epub 2017 Jul 19. Clin Cancer Res. 2017. PMID: 28724664 Free PMC article. - Fundamental concepts in genetics: genetics and the understanding of selection.
Hurst LD. Hurst LD. Nat Rev Genet. 2009 Feb;10(2):83-93. doi: 10.1038/nrg2506. Nat Rev Genet. 2009. PMID: 19119264 Review.
References
- Scheffler IE. Molecular genetics of succinate:quinone oxidoreductase in eukaryotes. Prog Nucleic Acid Res Mol Biol. 1998;60:267–315. - PubMed
- Baysal BE, Ferrell RE, Wilett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PEM, Rubinstein WS, Myers EN, Richard CW, III, Cornelisse CJ, Devilee P, Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous