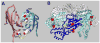Cellular functions of NSF: not just SNAPs and SNAREs - PubMed (original) (raw)
Review
Cellular functions of NSF: not just SNAPs and SNAREs
Chunxia Zhao et al. FEBS Lett. 2007.
Abstract
N-ethylmaleimide sensitive factor (NSF) is an ATPases associated with various cellular activities protein (AAA), broadly required for intracellular membrane fusion. NSF functions as a SNAP receptor (SNARE) chaperone which binds, through soluble NSF attachment proteins (SNAPs), to SNARE complexes and utilizes the energy of ATP hydrolysis to disassemble them thus facilitating SNARE recycling. While this is a major function of NSF, it does seem to interact with other proteins, such as the AMPA receptor subunit, GluR2, and beta2-AR and is thought to affect their trafficking patterns. New data suggest that NSF may be regulated by transient post-translational modifications such as phosphorylation and nitrosylation. These new aspects of NSF function as well as its role in SNARE complex dynamics will be discussed.
Figures
Figure 1. 20S Particle
Depicted are the crystal structures for each element of the 20S particle. The two ATP-binding domains of NSF (D1 and D2) in white are modeled from the NSF-D2 structure (1D2N). A trimer of NSF-N domains, in red, is based on the three-in-three-out model of May et al. 1999 (1QDN). Only two yeast α-SNAPs (Sec17p, 1QQE ) are depicted in yellow. The coiled-coil SNARE complex is depicted with syntaxin-1a in light blue, synaptobrevin/VAMP-2, in magenta, and SNAP-25 in dark blue (2BUO). The images were created with Swiss PDB viewer and rendered with Pov-Ray.
Figure 2. Post-translational modifications of NSF
Panel A, crystal structure of the amino-terminal domain of NSF, showing the locations of the modified cysteine and tyrosine residues. The NA subdomain is in rose and the NB subdomain is in aqua. The image is based on 1QDN and was generated using Swiss PDB viewer and rendered with Pov-Ray. Panel B, crystal structure of the NSF-D2 hexamer. Three subunits are shown in white, two in aqua and one in blue. The modified residues are indicated by red. Position 1 is predicted to be equivalent to the phosphorylated Ser237 in the NSF-D1 domain. Position 2 is Ser569, phosphorylated by Pctaire1. The image is based on 1D2N and was created with Swiss PDB viewer and rendered with Pov-Ray.
Similar articles
- Requirements for the catalytic cycle of the N-ethylmaleimide-Sensitive Factor (NSF).
Zhao C, Smith EC, Whiteheart SW. Zhao C, et al. Biochim Biophys Acta. 2012 Jan;1823(1):159-71. doi: 10.1016/j.bbamcr.2011.06.003. Epub 2011 Jun 13. Biochim Biophys Acta. 2012. PMID: 21689688 Free PMC article. Review. - Disassembly of all SNARE complexes by N-ethylmaleimide-sensitive factor (NSF) is initiated by a conserved 1:1 interaction between α-soluble NSF attachment protein (SNAP) and SNARE complex.
Vivona S, Cipriano DJ, O'Leary S, Li YH, Fenn TD, Brunger AT. Vivona S, et al. J Biol Chem. 2013 Aug 23;288(34):24984-91. doi: 10.1074/jbc.M113.489807. Epub 2013 Jul 8. J Biol Chem. 2013. PMID: 23836889 Free PMC article. - Mechanistic insights into the recycling machine of the SNARE complex.
Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Zhao M, et al. Nature. 2015 Feb 5;518(7537):61-7. doi: 10.1038/nature14148. Epub 2015 Jan 12. Nature. 2015. PMID: 25581794 Free PMC article. - Dissecting the N-ethylmaleimide-sensitive factor: required elements of the N and D1 domains.
Zhao C, Matveeva EA, Ren Q, Whiteheart SW. Zhao C, et al. J Biol Chem. 2010 Jan 1;285(1):761-72. doi: 10.1074/jbc.M109.056739. Epub 2009 Nov 3. J Biol Chem. 2010. PMID: 19887446 Free PMC article. - Recent Advances in Deciphering the Structure and Molecular Mechanism of the AAA+ ATPase N-Ethylmaleimide-Sensitive Factor (NSF).
Zhao M, Brunger AT. Zhao M, et al. J Mol Biol. 2016 May 8;428(9 Pt B):1912-26. doi: 10.1016/j.jmb.2015.10.026. Epub 2015 Nov 3. J Mol Biol. 2016. PMID: 26546278 Free PMC article. Review.
Cited by
- Identification of Novel Adenylyl Cyclase 5 (AC5) Signaling Networks in D1 and D2 Medium Spiny Neurons using Bimolecular Fluorescence Complementation Screening.
Doyle TB, Muntean BS, Ejendal KF, Hayes MP, Soto-Velasquez M, Martemyanov KA, Dessauer CW, Hu CD, Watts VJ. Doyle TB, et al. Cells. 2019 Nov 19;8(11):1468. doi: 10.3390/cells8111468. Cells. 2019. PMID: 31752385 Free PMC article. - Zipcode RNA-Binding Proteins and Membrane Trafficking Proteins Cooperate to Transport Glutelin mRNAs in Rice Endosperm.
Tian L, Doroshenk KA, Zhang L, Fukuda M, Washida H, Kumamaru T, Okita T. Tian L, et al. Plant Cell. 2020 Aug;32(8):2566-2581. doi: 10.1105/tpc.20.00111. Epub 2020 May 29. Plant Cell. 2020. PMID: 32471860 Free PMC article. - Requirements for the catalytic cycle of the N-ethylmaleimide-Sensitive Factor (NSF).
Zhao C, Smith EC, Whiteheart SW. Zhao C, et al. Biochim Biophys Acta. 2012 Jan;1823(1):159-71. doi: 10.1016/j.bbamcr.2011.06.003. Epub 2011 Jun 13. Biochim Biophys Acta. 2012. PMID: 21689688 Free PMC article. Review. - N-ethylmaleimide-sensitive factor interacts with the serotonin transporter and modulates its trafficking: implications for pathophysiology in autism.
Iwata K, Matsuzaki H, Tachibana T, Ohno K, Yoshimura S, Takamura H, Yamada K, Matsuzaki S, Nakamura K, Tsuchiya KJ, Matsumoto K, Tsujii M, Sugiyama T, Katayama T, Mori N. Iwata K, et al. Mol Autism. 2014 May 10;5:33. doi: 10.1186/2040-2392-5-33. eCollection 2014. Mol Autism. 2014. PMID: 24834316 Free PMC article. - The SNARE complex in neuronal and sensory cells.
Ramakrishnan NA, Drescher MJ, Drescher DG. Ramakrishnan NA, et al. Mol Cell Neurosci. 2012 May;50(1):58-69. doi: 10.1016/j.mcn.2012.03.009. Epub 2012 Apr 2. Mol Cell Neurosci. 2012. PMID: 22498053 Free PMC article. Review.
References
- Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–21. - PubMed
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–24. - PubMed
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–72. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials