Lifespan regulation by evolutionarily conserved genes essential for viability - PubMed (original) (raw)
Lifespan regulation by evolutionarily conserved genes essential for viability
Sean P Curran et al. PLoS Genet. 2007.
Abstract
Evolutionarily conserved mechanisms that control aging are predicted to have prereproductive functions in order to be subject to natural selection. Genes that are essential for growth and development are highly conserved in evolution, but their role in longevity has not previously been assessed. We screened 2,700 genes essential for Caenorhabditis elegans development and identified 64 genes that extend lifespan when inactivated postdevelopmentally. These candidate lifespan regulators are highly conserved from yeast to humans. Classification of the candidate lifespan regulators into functional groups identified the expected insulin and metabolic pathways but also revealed enrichment for translation, RNA, and chromatin factors. Many of these essential gene inactivations extend lifespan as much as the strongest known regulators of aging. Early gene inactivations of these essential genes caused growth arrest at larval stages, and some of these arrested animals live much longer than wild-type adults. daf-16 is required for the enhanced survival of arrested larvae, suggesting that the increased longevity is a physiological response to the essential gene inactivation. These results suggest that insulin-signaling pathways play a role in regulation of aging at any stage in life.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
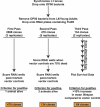
Figure 1. Representation of the Postdevelopmental RNAi Screen for Longevity
Embryos from gravid C. elegans hermaphrodites were isolated by hypochlorite treatment and allowed to hatch overnight in the absence of food. The synchronized L1 larvae were then placed on OP50 agar plates at 20 °C and allowed to develop to L4/young adult stage. The L4/young adult animals were then washed from the plates, cleaned from bacteria by sucrose flotation, and placed on six-well RNAi plates (∼30 worms/well). From three replicates of the first pass, 470 RNAi clones were identified as positive and were retested in the second pass under similar conditions while increasing the stringency for scoring positive. A total of 134 RNAi clones passed the second pass criteria and were scored longitudinally in the third pass against blind positive (daf-2 RNAi) and negative (empty vector, eri-1 RNAi, and daf-16 RNAi) controls. A total of 64 RNAi clones increased mean lifespan by >10% compared to the negative controls.
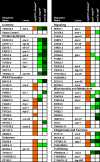
Figure 2. Identification of RNAi Clones that Act through DAF-16
(A) Shading indicates that daf-16(mgDf47); eri-1(mg366) is epistatic to the lifespan extension observed in eri-1(mg366) (Table S2). (B) DAF-16 nuclear localization after feeding RNAi clone is shown. Light shading indicates some nuclear localization, and medium shading indicates more nuclear localization (Figure S4). (C) sod-3p::gfp expression after RNAi feeding is shown. Light shading indicates weak expression, medium shading indicated modest expression, and dark shading indicates strong expression.
Similar articles
- The zinc-finger protein SEA-2 regulates larval developmental timing and adult lifespan in C. elegans.
Huang X, Zhang H, Zhang H. Huang X, et al. Development. 2011 May;138(10):2059-68. doi: 10.1242/dev.057109. Epub 2011 Apr 6. Development. 2011. PMID: 21471153 - DAF-16 target genes that control C. elegans life-span and metabolism.
Lee SS, Kennedy S, Tolonen AC, Ruvkun G. Lee SS, et al. Science. 2003 Apr 25;300(5619):644-7. doi: 10.1126/science.1083614. Epub 2003 Apr 10. Science. 2003. PMID: 12690206 - Longevity determined by developmental arrest genes in Caenorhabditis elegans.
Chen D, Pan KZ, Palter JE, Kapahi P. Chen D, et al. Aging Cell. 2007 Aug;6(4):525-33. doi: 10.1111/j.1474-9726.2007.00305.x. Epub 2007 May 29. Aging Cell. 2007. PMID: 17521386 Free PMC article. - The search for DAF-16/FOXO transcriptional targets: approaches and discoveries.
Murphy CT. Murphy CT. Exp Gerontol. 2006 Oct;41(10):910-21. doi: 10.1016/j.exger.2006.06.040. Epub 2006 Aug 24. Exp Gerontol. 2006. PMID: 16934425 Review. - Genetic control of longevity in C. elegans.
Braeckman BP, Vanfleteren JR. Braeckman BP, et al. Exp Gerontol. 2007 Jan-Feb;42(1-2):90-8. doi: 10.1016/j.exger.2006.04.010. Epub 2006 Jul 10. Exp Gerontol. 2007. PMID: 16829009 Review.
Cited by
- A cytoprotective perspective on longevity regulation.
Shore DE, Ruvkun G. Shore DE, et al. Trends Cell Biol. 2013 Sep;23(9):409-20. doi: 10.1016/j.tcb.2013.04.007. Epub 2013 May 30. Trends Cell Biol. 2013. PMID: 23726168 Free PMC article. Review. - APL-1, the Alzheimer's Amyloid precursor protein in Caenorhabditis elegans, modulates multiple metabolic pathways throughout development.
Ewald CY, Raps DA, Li C. Ewald CY, et al. Genetics. 2012 Jun;191(2):493-507. doi: 10.1534/genetics.112.138768. Epub 2012 Mar 30. Genetics. 2012. PMID: 22466039 Free PMC article. - Using C. elegans for aging research.
Tissenbaum HA. Tissenbaum HA. Invertebr Reprod Dev. 2015 Jan 30;59(sup1):59-63. doi: 10.1080/07924259.2014.940470. Epub 2014 Dec 9. Invertebr Reprod Dev. 2015. PMID: 26136622 Free PMC article. - An automated microfluidic platform for C. elegans embryo arraying, phenotyping, and long-term live imaging.
Cornaglia M, Mouchiroud L, Marette A, Narasimhan S, Lehnert T, Jovaisaite V, Auwerx J, Gijs MA. Cornaglia M, et al. Sci Rep. 2015 May 7;5:10192. doi: 10.1038/srep10192. Sci Rep. 2015. PMID: 25950235 Free PMC article. - Considering Caenorhabditis elegans Aging on a Temporal and Tissue Scale: The Case of Insulin/IGF-1 Signaling.
Fabrizio P, Alcolei A, Solari F. Fabrizio P, et al. Cells. 2024 Feb 5;13(3):288. doi: 10.3390/cells13030288. Cells. 2024. PMID: 38334680 Free PMC article. Review.
References
- Finch CE, Ruvkun G. The genetics of aging. Annu Rev Genomics Hum Genet. 2001;2:435–462. - PubMed
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. - PubMed
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. - PubMed
- Kawano T, Kataoka N, Abe S, Ohtani M, Honda Y, et al. Lifespan extending activity of substances secreted by the nematode Caenorhabditis elegans that include the dauer-inducing pheromone. Biosci Biotechnol Biochem. 2005;69:2479–2481. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG016636/AG/NIA NIH HHS/United States
- F32 AG026207-02/AG/NIA NIH HHS/United States
- F32-AG026207/AG/NIA NIH HHS/United States
- R01-AG016636/AG/NIA NIH HHS/United States
- R00 AG032308/AG/NIA NIH HHS/United States
- F32 AG026207/AG/NIA NIH HHS/United States
- F32 AG026207-03/AG/NIA NIH HHS/United States
- F32 AG026207-01/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous