C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing - PubMed (original) (raw)
C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing
Geurt Schilders et al. Nucleic Acids Res. 2007.
Abstract
The exosome is a complex of 3'-5' exoribonucleases and RNA-binding proteins, which is involved in processing or degradation of different classes of RNA. Previously, the characterization of purified exosome complexes from yeast and human cells suggested that C1D and KIAA0052/hMtr4p are associated with the exosome and thus might regulate its functional activities. Subcellular localization experiments demonstrated that C1D and KIAA0052/hMtr4p co-localize with exosome subunit PM/Scl-100 in the nucleoli of HEp-2 cells. Additionally, the nucleolar accumulation of C1D appeared to be dependent on PM/Scl-100. Protein-protein interaction studies showed that C1D binds to PM/Scl-100, whereas KIAA0052/hMtr4p was found to interact with MPP6, a previously identified exosome-associated protein. Moreover, we demonstrate that C1D, MPP6 and PM/Scl-100 form a stable trimeric complex in vitro. Knock-down of C1D, MPP6 and KIAA0052/hMtr4p by RNAi resulted in the accumulation of 3'-extended 5.8S rRNA precursors, showing that these proteins are required for rRNA processing. Interestingly, C1D appeared to contain RNA-binding activity with a potential preference for structured RNAs. Taken together, our results are consistent with a role for the exosome-associated proteins C1D, MPP6 and KIAA052/hMtr4p in the recruitment of the exosome to pre-rRNA to mediate the 3' end processing of the 5.8S rRNA.
Figures
Figure 1.
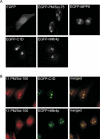
C1D and hMtr4p co-localize with PM/Scl-100 in the nucleoli of HEp-2 cells. (A) HEp-2 cells were transiently transfected with cDNA constructs encoding EGFP alone, EGFP-PM/Scl-75, EGFP-MPP6, EGFP-C1D or EGFP-hMtr4p. Twenty-four hours after transfection, the cells were fixed and EGFP or EGFP-fusion proteins were visualized by confocal fluorescence microscopy. (B) HEp-2 cells were transfected with cDNA constructs encoding C1D and hMtr4p fused to the C-terminus of EGFP; 24 h after transfection, cells were fixed and incubated with rabbit anti-PM/Scl-100 antibodies, which were visualized by Texas-Red-conjugated secondary antibodies (left panels). The localization of EGFP-tagged C1D and hMtr4p is shown in the middle panels, and the corresponding merged images are shown on the right.
Figure 2.
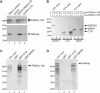
Association of C1D, MPP6 and hMtr4p with components of the exosome. (A) Co-immunoprecipitations were performed using anti-EGFP antibodies and extracts of HEp-2 cells transiently transfected with expression constructs for either EGFP alone (lane 2), EGFP-PM/Scl-75 (lane 3) or EGFP-C1D (lane 4). In the first lane, total extract from non-transfected HEp-2 cells was separated. The precipitated proteins were analyzed by western blotting, using anti-PM/Scl-positive patient serum R212 (upper part of the blot) or a monoclonal antibody to hRrp4p (lower part of the blot). Arrows indicate the positions of PM/Scl-100 and hRrp4p. The positions of molecular weight markers are indicated on the left. (B) Bacterially expressed, recombinant His-tagged PM/Scl-100 was immobilized using anti-PM/Scl-100 antibodies and incubated with either 35S-labeled, in vitro translated GAPDH (lane 1), C1D (lane 4) or MPP6 (lane 7). Co-precipitated proteins were analyzed by SDS-PAGE and autoradiography. The positions of these proteins are indicated with arrows, and the positions of molecular weight markers are indicated on the left. Lanes 3, 6 and 9 show proteins co-precipitated with His-PM/Scl-100. In lanes 2, 5 and 8, material from control incubations, in which no recombinant His-PM/Scl-100 was added, was analyzed. (C) Glutathione-Sepharose beads were used to precipitate GST (lane 2), GST-C1D (lane 3) or GST-MPP6 (lane 4), which were incubated with 35S-labeled, in vitro translated PM/Scl-100. After precipitation, bound PM/Scl-100 was analyzed by SDS-PAGE and autoradiography. In lane 1, 10% of the amount of labeled PM/Scl-100 used per incubation was loaded. On the left, the positions of molecular weight markers are indicated. (D) Similar experiments as described in (C), but now with 35S-labeled, in vitro translated hMtr4p instead of PM/Scl-100.
Figure 3.
C1D, MPP6 and PM/Scl-100 form a trimeric complex in vitro. GST-tagged C1D was incubated with either 35S-labeled, in vitro translated C1D (lane 5), MPP6 (lane 6), PM/Scl-100 (lane 7) or MPP6 and PM/Scl-100 (lane 8). As a control, GST alone was incubated with all three labeled proteins (lane 4). After incubation, GST(-C1D)-containing complexes were precipitated with glutathione-Sepharose beads and analyzed by SDS-PAGE and autoradiography. In lanes 1–3, the in vitro translated C1D, MPP6 and PM/Scl-100 proteins were analyzed.
Figure 4.
Nucleolar accumulation of C1D requires PM/Scl-100. HEp-2 cells were transfected with a construct encoding C1D fused to the N-terminus of EGFP and 16 h after transfection cells were treated with a control siRNA (lanes 1, 3 and 5), or siRNAs targeting PM/Scl-75 (lane 2), MPP6 (lane 4) or PM/Scl-100 (lane 6). (A) After 24 h, cells were harvested, and total cell extracts were analyzed by western blotting using a polyclonal anti-PM/Scl-75 serum (lanes 1 and 2), a polyclonal anti-MPP6 serum (lanes 3 and 4) and a polyclonal anti-PM/Scl-100 serum (lanes 5 and 6). A mouse monoclonal antibody to gamma tubulin was used as a control. (B) Twenty-four hours after siRNA treatment, cells were fixed and the expressed C1D-EGFP fusion protein was visualized by fluorescence microscopy. Panel control siRNA, cells treated with a control siRNA; panel PM/Scl-75 siRNA, cells treated with the siRNA targeting PM/Scl-75; panel MPP6 siRNA, cells treated with the siRNA targeting MPP6; panel PM/Scl-100 siRNA, cells treated with the siRNA targeting PM/Scl-100. (C) To investigate the integrity of the C1D-EGFP fusion protein upon PM/Scl-100 siRNA-mediated knock-down, extracts were prepared from the transfected HEp-2 cells and analyzed by western blotting using a polyclonal anti-EGFP antiserum. A mouse monoclonal antibody directed against gamma tubulin was used as a control.
Figure 5.
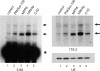
Knock-down of hMtr4p and C1D leads to the accumulation of 5.8S rRNA precursors. HEp-2 cells were transiently transfected with siRNAs directed to PM/Scl-100 (lane 2), MPP6 (lane 3), hMtr4p (lane 4), C1D (lane 5) or with a control siRNA (lane 1). Cells were harvested 2 days after transfection and 2.5 µg of total RNA was analyzed by northern blot hybridization using radiolabeled probes specific for 5.8S rRNA (left) or ITS2 (upper right). As a control, a U6 snRNA probe was used (lower right). The positions of the 5.8S rRNA precursors are indicated by arrows and the position of mature 5.8S rRNA is indicated with an asterisk.
Figure 6.
C1D binds RNA in vitro. GST and GST-C1D were incubated with radiolabeled homopolynucleotides and bound RNAs were quantified in a scintillation counter. The binding efficiency of GST and GST-MPP6 to poly(A), poly(C), poly(G), poly(U) and poly(I)–poly(C) is depicted as a percentage of input RNA (RBE: relative binding efficiency). These results are the average values of two independent experiments.
Similar articles
- MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation.
Schilders G, Raijmakers R, Raats JM, Pruijn GJ. Schilders G, et al. Nucleic Acids Res. 2005 Dec 7;33(21):6795-804. doi: 10.1093/nar/gki982. Print 2005. Nucleic Acids Res. 2005. PMID: 16396833 Free PMC article. - NVL2, a nucleolar AAA-ATPase, is associated with the nuclear exosome and is involved in pre-rRNA processing.
Yoshikatsu Y, Ishida Y, Sudo H, Yuasa K, Tsuji A, Nagahama M. Yoshikatsu Y, et al. Biochem Biophys Res Commun. 2015 Aug 28;464(3):780-6. doi: 10.1016/j.bbrc.2015.07.032. Epub 2015 Jul 10. Biochem Biophys Res Commun. 2015. PMID: 26166824 - Interaction profiling identifies the human nuclear exosome targeting complex.
Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Lubas M, et al. Mol Cell. 2011 Aug 19;43(4):624-37. doi: 10.1016/j.molcel.2011.06.028. Mol Cell. 2011. PMID: 21855801 - The human exosome: an autoantigenic complex of exoribonucleases in myositis and scleroderma.
Brouwer R, Pruijn GJ, van Venrooij WJ. Brouwer R, et al. Arthritis Res. 2001;3(2):102-6. doi: 10.1186/ar147. Epub 2000 Dec 20. Arthritis Res. 2001. PMID: 11178117 Free PMC article. Review. - Rrp6, Rrp47 and cofactors of the nuclear exosome.
Butler JS, Mitchell P. Butler JS, et al. Adv Exp Med Biol. 2010;702:91-104. Adv Exp Med Biol. 2010. PMID: 21618877 Review.
Cited by
- Intronless mRNAs transit through nuclear speckles to gain export competence.
Wang K, Wang L, Wang J, Chen S, Shi M, Cheng H. Wang K, et al. J Cell Biol. 2018 Nov 5;217(11):3912-3929. doi: 10.1083/jcb.201801184. Epub 2018 Sep 7. J Cell Biol. 2018. PMID: 30194269 Free PMC article. - Differential expression of RNA exosome subunits in the amphibian Lithobates catesbeianus during reproductive and non-reproductive periods.
Luz JS, Caneguim BH, Baggio A, Santoni MM, Helbing CC, Valentini SR, Sasso-Cerri E, Oliveira CC. Luz JS, et al. BMC Res Notes. 2019 Jan 21;12(1):46. doi: 10.1186/s13104-019-4077-7. BMC Res Notes. 2019. PMID: 30665462 Free PMC article. - The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2.
Schillewaert S, Wacheul L, Lhomme F, Lafontaine DL. Schillewaert S, et al. Mol Cell Biol. 2012 Jan;32(2):430-44. doi: 10.1128/MCB.06019-11. Epub 2011 Nov 14. Mol Cell Biol. 2012. PMID: 22083961 Free PMC article. - An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression.
Ogami K, Richard P, Chen Y, Hoque M, Li W, Moresco JJ, Yates JR 3rd, Tian B, Manley JL. Ogami K, et al. Genes Dev. 2017 Jun 15;31(12):1257-1271. doi: 10.1101/gad.302604.117. Epub 2017 Jul 21. Genes Dev. 2017. PMID: 28733371 Free PMC article. - Progenitor function in self-renewing human epidermis is maintained by the exosome.
Mistry DS, Chen Y, Sen GL. Mistry DS, et al. Cell Stem Cell. 2012 Jul 6;11(1):127-35. doi: 10.1016/j.stem.2012.04.022. Cell Stem Cell. 2012. PMID: 22770246 Free PMC article.
References
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. - PubMed
- Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous