The innate immune repertoire in cnidaria--ancestral complexity and stochastic gene loss - PubMed (original) (raw)
Comparative Study
The innate immune repertoire in cnidaria--ancestral complexity and stochastic gene loss
David J Miller et al. Genome Biol. 2007.
Abstract
Background: Characterization of the innate immune repertoire of extant cnidarians is of both fundamental and applied interest--it not only provides insights into the basic immunological 'tool kit' of the common ancestor of all animals, but is also likely to be important in understanding the global decline of coral reefs that is presently occurring. Recently, whole genome sequences became available for two cnidarians, Hydra magnipapillata and Nematostella vectensis, and large expressed sequence tag (EST) datasets are available for these and for the coral Acropora millepora.
Results: To better understand the basis of innate immunity in cnidarians, we scanned the available EST and genomic resources for some of the key components of the vertebrate innate immune repertoire, focusing on the Toll/Toll-like receptor (TLR) and complement pathways. A canonical Toll/TLR pathway is present in representatives of the basal cnidarian class Anthozoa, but neither a classic Toll/TLR receptor nor a conventional nuclear factor (NF)-kappaB could be identified in the anthozoan Hydra. Moreover, the detection of complement C3 and several membrane attack complex/perforin domain (MAC/PF) proteins suggests that a prototypic complement effector pathway may exist in anthozoans, but not in hydrozoans. Together with data for several other gene families, this implies that Hydra may have undergone substantial secondary gene loss during evolution. Such losses are not confined to Hydra, however, and at least one MAC/PF gene appears to have been lost from Nematostella.
Conclusion: Consideration of these patterns of gene distribution underscores the likely significance of gene loss during animal evolution whilst indicating ancient origins for many components of the vertebrate innate immune system.
Figures
Figure 1
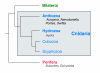
Relationships at the base of the Metazoa. Cnidarians are amongst the simplest animals at the tissue grade of organization, and are often regarded as the closest outgroup to the Bilateria. Within the Cnidaria, the class Anthozoa is basal, whereas the Hydrozoa is derived. The sponges (Porifera) are unquestionably animals, but represent a lower level of organization. The affinities and relationships of genera mentioned in the text are indicated.
Figure 2
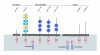
Summary of domain structures of TIR domain-containing proteins identified in selected Cnidaria.
Figure 3
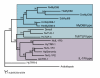
Phylogenetic analysis of cnidarian TIR sequences in comparison to a selection of TIR domains from other species. The maximum likelihood (ML) tree shown is the result of analysis of an HMM-based alignment of TIR domains. A number of the TIR sequences identified and discussed in the text are incomplete due to the presence of introns of unknown size, and hence were not included in the phylogenetic analyses. Three clades are resolved by these analyses, corresponding to the TIR domains characteristic of the 'MyD88-type', 'Toll/TLR-type' and 'IL-1R-type'. In addition to the TIR domain, the first of these types contains a death domain and the second contains multiple LRRs. Like the mammalian receptors for interleukin 1, the three Nematostella proteins falling into the third clade each also contain multiple immunoglobulin domains. Note that HyTRR1 does not contain such domains and that it is not yet clear whether either of the Acropora proteins does. The Acropora sequences included in the analysis were predicted from A. palmata genomic clones (ApGenomic) and from an A. millepora cDNA clone (AmTIR-1). Hydra lacks a canonical Toll/TLR, having only two MyD88 genes and the two sequences known as TRR-1 and TRR-2; H. magnipapillata and N. vectensis sequences are indicated by the prefixes Hy and Nv, respectively. Reference sequences: HsMyD88, human MyD88 (SwissProt:Q99836); DmMyD88, fly MyD88 (GenBank:AAL56570); SdMyD88, Suberites MyD88 (EMBL:CAI68016); Dmtoll, fly Toll (SwissProt:P08953); HsTLR4, human TLR4 (EMBL:CAD99157); Arabidopsis (GenBank:AAN28912).
Figure 4
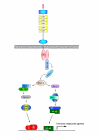
Signaling pathways downstream of the Toll/TLRs. Pattern recognition, either indirectly or directly, by Toll/TLRs results in activation of NF-κB (vertebrates) or the Dif/Rel heterodimer (Drosophila) and thus transcription of appropriate immune response genes. At TRAF6, the classic Toll/TIR pathway (shown in the right branch) is linked to the JNK/p38 pathway (shown in the left branch) by the ECSIT protein, which acts as a regulator of MEKK-1 processing [35]. Components of both pathways downstream of Toll/TLRs are represented in the cnidarian datasets (Table 1). ECSIT may also act as a link between these and the TGF-b signaling pathway, since it forms complexes with BMP-pathway restricted Smads and is essential for regulation of the BMP-target gene Tlx2 [36]. All of the components of the TGF-b signaling pathway are also known from anthozoan cnidarians [13].
Figure 5
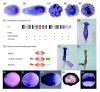
Complement component C3 and MAC/PF domain-containing proteins in Cnidaria. (a-e) In situ hybridization of C3-Am in Acropora. Expression first becomes apparent in scattered endodermal cells concentrated at the aboral end as the planula elongates from sphere to pear (a) and eventually to spindle (b). Endodermal expression continues post-settlement (c-e), becoming especially strong in the upper part of the polyp as it rises from the calcifying base (d). Post-settlement, the polyp consists of a series of hollow chambers interconnected beneath the mouth. The line of strong staining peripherally is the result of viewing the endoderm vertically, while elsewhere one is looking through the staining layer. (f) Domain map and presence (+)/absence (-) data for the various protein domains characteristic of complement C3 components in the Hydra, Nematostella and Acropora datasets. (g) In situ hybridization of the H. magnipapillata A2M-related gene. Hydra A2M-related transcripts are present in the endoderm along the whole body axis. Note that this Hydra gene lacks several of the C3-diagnostic domains that are present in the anthozoan C3s (see text). (h) Domain maps of major cnidarian MAC/PF proteins types. (i) Hydra Tx-60a in situ. The insert shows the sense control. (j) Hydra apextrin in situ. (k-o) Acropora apextrin in situ. Expression is first apparent in scattered ectodermal cells orally as the planula begins to elongate (k). At slightly later stages expression has spread toward the aboral end of the planula, still in scattered cells (l). As the elongation process continues, uniform strong expression is localized in all ectodermal cells in the oral two-thirds of the planula (m). The strong ectodermal expression is clearly apparent in this transilluminated transverse section cut from the central region of the planula (n). Following settlement, expression continues at the oral end of the planula, before frequently becoming limited to a narrow ring separating oral and aboral tissue (o).
Similar articles
- The evolution of immunity: a low-life perspective.
Hemmrich G, Miller DJ, Bosch TC. Hemmrich G, et al. Trends Immunol. 2007 Oct;28(10):449-54. doi: 10.1016/j.it.2007.08.003. Epub 2007 Sep 12. Trends Immunol. 2007. PMID: 17855167 - Domain combination of the vertebrate-like TLR gene family: implications for their origin and evolution.
Wu B, Huan T, Gong J, Zhou P, Bai Z. Wu B, et al. J Genet. 2011 Dec;90(3):401-8. doi: 10.1007/s12041-011-0097-3. J Genet. 2011. PMID: 22227927 - Pax gene diversity in the basal cnidarian Acropora millepora (Cnidaria, Anthozoa): implications for the evolution of the Pax gene family.
Miller DJ, Hayward DC, Reece-Hoyes JS, Scholten I, Catmull J, Gehring WJ, Callaerts P, Larsen JE, Ball EE. Miller DJ, et al. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4475-80. doi: 10.1073/pnas.97.9.4475. Proc Natl Acad Sci U S A. 2000. PMID: 10781047 Free PMC article. - Recent advances in genomics and transcriptomics of cnidarians.
Technau U, Schwaiger M. Technau U, et al. Mar Genomics. 2015 Dec;24 Pt 2:131-8. doi: 10.1016/j.margen.2015.09.007. Epub 2015 Oct 1. Mar Genomics. 2015. PMID: 26421490 Review. - The Complicated Evolutionary Diversification of the Mpeg-1/Perforin-2 Family in Cnidarians.
Walters BM, Connelly MT, Young B, Traylor-Knowles N. Walters BM, et al. Front Immunol. 2020 Aug 6;11:1690. doi: 10.3389/fimmu.2020.01690. eCollection 2020. Front Immunol. 2020. PMID: 32849589 Free PMC article. Review.
Cited by
- Innate immunity in the simplest animals - placozoans.
Kamm K, Schierwater B, DeSalle R. Kamm K, et al. BMC Genomics. 2019 Jan 5;20(1):5. doi: 10.1186/s12864-018-5377-3. BMC Genomics. 2019. PMID: 30611207 Free PMC article. - Differential gene expression analysis of symbiotic and aposymbiotic Exaiptasia anemones under immune challenge with Vibrio coralliilyticus.
Roesel CL, Vollmer SV. Roesel CL, et al. Ecol Evol. 2019 Jun 30;9(14):8279-8293. doi: 10.1002/ece3.5403. eCollection 2019 Jul. Ecol Evol. 2019. PMID: 31380089 Free PMC article. - Microarray analysis identifies candidate genes for key roles in coral development.
Grasso LC, Maindonald J, Rudd S, Hayward DC, Saint R, Miller DJ, Ball EE. Grasso LC, et al. BMC Genomics. 2008 Nov 14;9:540. doi: 10.1186/1471-2164-9-540. BMC Genomics. 2008. PMID: 19014561 Free PMC article. - A novel gene family controls species-specific morphological traits in Hydra.
Khalturin K, Anton-Erxleben F, Sassmann S, Wittlieb J, Hemmrich G, Bosch TC. Khalturin K, et al. PLoS Biol. 2008 Nov 18;6(11):e278. doi: 10.1371/journal.pbio.0060278. PLoS Biol. 2008. PMID: 19018660 Free PMC article. - The presence of biomarker enzymes of selected Scleractinian corals of Palk Bay, southeast coast of India.
Anithajothi R, Duraikannu K, Umagowsalya G, Ramakritinan CM. Anithajothi R, et al. Biomed Res Int. 2014;2014:684874. doi: 10.1155/2014/684874. Epub 2014 Aug 18. Biomed Res Int. 2014. PMID: 25215288 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous