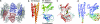Normal mode refinement of anisotropic thermal parameters for a supramolecular complex at 3.42-A crystallographic resolution - PubMed (original) (raw)
Normal mode refinement of anisotropic thermal parameters for a supramolecular complex at 3.42-A crystallographic resolution
Billy K Poon et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Here we report a normal-mode-based protocol for modeling anisotropic thermal motions of proteins in x-ray crystallographic refinement. The foundation for this protocol is a recently developed elastic normal mode analysis that produces much more accurate eigenvectors without the tip effect. The effectiveness of the procedure is demonstrated on the refinement of a 3.42-A structure of formiminotransferase cyclodeaminase, a 0.5-MDa homooctameric enzyme. Using an order of magnitude fewer adjustable thermal parameters than the conventional isotropic refinement, this protocol resulted in a decrease of the values of R(cryst) and R(free) and improvements of the density map. Several poorly resolved regions in the original isotropically refined structure became clearer so that missing side chains were fitted easily and mistraced backbone was corrected. Moreover, the distribution of anisotropic thermal ellipsoids revealed functionally important structure flexibility. This normal-mode-based refinement is an effective way of describing anisotropic thermal motions in x-ray structures and is particularly attractive for the refinement of very large and flexible supramolecular complexes at moderate resolutions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Structure of FTCD. (a) The square doughnut structure of an FTCD octamer. Two subunits are shown in red and blue, respectively. (b) The subunit structure of ligand-free FTCD. Backbone trace color ramped from the N terminus to the C terminus. (c) Superposition of the FT domain of human ligand-free FTCD (red) with the structure of the same domain in isolation (cyan) with the product analog, folinic acid (CPK mode), bound in the groove. (d) Rainbow-colored isotropic B factor in the original model. The hotter the color, the larger the B factors. The high flexibility of the N-subdomain, the linker region, and the lower half of the CD domain is evident.
Fig. 2.
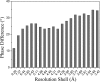
Phase shifts of the normal-mode-refined model with respect to the original model. The results are shown as a function of resolution shells. Larger phase shifts are observed for higher resolution shells. Data were calculated by the SFTOOLS program in CCP4 suite.
Fig. 3.
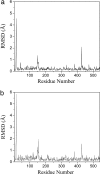
Structural shifts of the normal-mode-refined model with respect to the original model. The rmsd (angstroms) along the chain of a single subunit is shown for both main chains (a) and side chains (b). Three large spikes are evident in both graphs. The results for other subunits are very similar because of a noncrystallographic symmetry constraint.
Fig. 4.
Examples of large structural adjustments in normal-mode refinement. (Upper) The original models. (Lower) Normal-mode model. (a and a′) Region Glu-13–Asn-15 superimposed with omit 2_F_o − _F_c map contoured at 1.5σ. (b and b′) Region Glu-147–Pro-150 superimposed with omit 2_F_o − _F_c map contoured at 1.0σ. In both b and b′, the original model (pink) and the new model (yellow) are superimposed to highlight the structural shifts. (c and c′) Region Pro-426–Lys-427 superimposed with omit 2_F_o − _F_c map contoured at 1.0σ. (d and d′) Residue Se-Met-132 superimposed with omit 2_F_o − _F_c map contoured at 1.5σ.
Fig. 5.
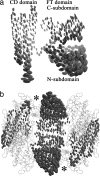
Anisotropic deformations of FTCD revealed by the normal-mode refinement. (a) Thermal ellipsoids for a single subunit of FTCD in the same view as Fig. 1_b_. It is evident that the N-subdomain of the FT domain and the lower half of the CD domain are highly flexible. (b) Side view of the square doughnut of octameric FTCD. For clarity, only the ellipsoids of two FT domains and two CD domains in a neighboring subunit are shown. The two catalytic reaction centers are indicated by the asterisks.
Similar articles
- Normal-mode refinement of anisotropic thermal parameters for potassium channel KcsA at 3.2 A crystallographic resolution.
Chen X, Poon BK, Dousis A, Wang Q, Ma J. Chen X, et al. Structure. 2007 Aug;15(8):955-62. doi: 10.1016/j.str.2007.06.012. Structure. 2007. PMID: 17698000 Free PMC article. - Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement.
Chen X, Lu M, Poon BK, Wang Q, Ma J. Chen X, et al. Acta Crystallogr D Biol Crystallogr. 2009 Apr;65(Pt 4):339-47. doi: 10.1107/S0907444909003539. Epub 2009 Mar 19. Acta Crystallogr D Biol Crystallogr. 2009. PMID: 19307715 Free PMC article. - Anisotropic refinement of the structure of Thermoascus aurantiacus xylanase I.
Teixeira S, Lo Leggio L, Pickersgill R, Cardin C. Teixeira S, et al. Acta Crystallogr D Biol Crystallogr. 2001 Mar;57(Pt 3):385-92. doi: 10.1107/s0907444900019089. Acta Crystallogr D Biol Crystallogr. 2001. PMID: 11223515 - Expanding the model: anisotropic displacement parameters in protein structure refinement.
Merritt EA. Merritt EA. Acta Crystallogr D Biol Crystallogr. 1999 Jun;55(Pt 6):1109-17. doi: 10.1107/s0907444999003789. Acta Crystallogr D Biol Crystallogr. 1999. PMID: 10329772 Review. - Coarse-grained normal mode analysis in structural biology.
Bahar I, Rader AJ. Bahar I, et al. Curr Opin Struct Biol. 2005 Oct;15(5):586-92. doi: 10.1016/j.sbi.2005.08.007. Curr Opin Struct Biol. 2005. PMID: 16143512 Free PMC article. Review.
Cited by
- Structural basis of actin filament nucleation by tandem W domains.
Chen X, Ni F, Tian X, Kondrashkina E, Wang Q, Ma J. Chen X, et al. Cell Rep. 2013 Jun 27;3(6):1910-20. doi: 10.1016/j.celrep.2013.04.028. Epub 2013 May 30. Cell Rep. 2013. PMID: 23727244 Free PMC article. - Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells.
Chen X, Ni F, Kondrashkina E, Ma J, Wang Q. Chen X, et al. Proc Natl Acad Sci U S A. 2015 Oct 13;112(41):12687-92. doi: 10.1073/pnas.1512464112. Epub 2015 Sep 28. Proc Natl Acad Sci U S A. 2015. PMID: 26417072 Free PMC article. - A comparative analysis of the equilibrium dynamics of a designed protein inferred from NMR, X-ray, and computations.
Liu L, Koharudin LM, Gronenborn AM, Bahar I. Liu L, et al. Proteins. 2009 Dec;77(4):927-39. doi: 10.1002/prot.22518. Proteins. 2009. PMID: 19688820 Free PMC article. - Normal Mode Analysis as a Routine Part of a Structural Investigation.
Bauer JA, Pavlović J, Bauerová-Hlinková V. Bauer JA, et al. Molecules. 2019 Sep 10;24(18):3293. doi: 10.3390/molecules24183293. Molecules. 2019. PMID: 31510014 Free PMC article. Review. - Normal mode analysis with molecular geometry restraints: bridging molecular mechanics and elastic models.
Lu M, Ma J. Lu M, et al. Arch Biochem Biophys. 2011 Apr 1;508(1):64-71. doi: 10.1016/j.abb.2010.12.031. Epub 2011 Jan 4. Arch Biochem Biophys. 2011. PMID: 21211510 Free PMC article.
References
- Vitkup D, Ringe D, Karplus M, Petsko GA. Proteins. 2002;46:345–354. - PubMed
- Gerstein M, Lesk AM, Chothia C. Biochemistry. 1994;33:6739–6749. - PubMed
- Ma J. Structure (London) 2005;13:373–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM067801/GM/NIGMS NIH HHS/United States
- R01 GM068826/GM/NIGMS NIH HHS/United States
- R01-GM067801/GM/NIGMS NIH HHS/United States
- R01-GM068826/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials