TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer - PubMed (original) (raw)
TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer
Anita Langerød et al. Breast Cancer Res. 2007.
Abstract
Introduction: Gene expression profiling of breast carcinomas has increased our understanding of the heterogeneous biology of this disease and promises to impact clinical care. The aim of this study was to evaluate the prognostic value of gene expression-based classification along with established prognostic markers and mutation status of the TP53 gene (tumour protein p53) in a group of breast cancer patients with long-term (12 to 16 years) follow-up.
Methods: The clinical and histopathological parameters of 200 breast cancer patients were studied for their effects on clinical outcome using univariate/multivariate Cox regression. The prognostic impact of mutations in the TP53 gene, identified using temporal temperature gradient gel electrophoresis and sequencing, was also evaluated. Eighty of the samples were analyzed for gene expression using 42 K cDNA microarrays and the patients were assigned to five previously defined molecular expression groups. The strength of the gene expression based classification versus standard markers was evaluated by adding this variable to the Cox regression model used to analyze all samples.
Results: Both univariate and multivariate analysis showed that TP53 mutation status, tumor size and lymph node status were the strongest predictors of breast cancer survival for the whole group of patients. Analyses of the patients with gene expression data showed that TP53 mutation status, gene expression based classification, tumor size and lymph node status were significant predictors of survival. Breast cancer cases in the 'basal-like' and 'ERBB2+' gene expression subgroups had a very high mortality the first two years, while the 'highly proliferating luminal' cases developed the disease more slowly, showing highest mortality after 5 to 8 years. The TP53 mutation status showed strong association with the 'basal-like' and 'ERBB2+' subgroups, and tumors with mutation had a characteristic gene expression pattern.
Conclusion: TP53 mutation status and gene-expression based groups are important survival markers of breast cancer, and these molecular markers may provide prognostic information that complements clinical variables. The study adds experience and knowledge to an ongoing characterization and classification of the disease.
Figures
Figure 1
Hierarchical clustering using 'intrinsic' genes. (a) Five groups were identified, namely the highly proliferating luminal (light blue), luminal A (dark blue), normal-like (green), basal-like (red) and ERBB2+ (magenta) groups, which were used in the survival analysis. (b) Hierarchical clustering was performed using 540 clones, representing 496 unique genes from the 'intrinsic' gene list. The individual color of the dendrogram branches reflects the correlation with centroids previously described, and tumors with low correlation (<0.2) with a specific subtype are here indicated by gray branches. Gene clusters characterizing the five groups (a) involve genes encoding, for example, (c) estrogen receptor (ER), (d) MUC1, (e) cadherin 1 (E-cadherin; CDH1), (f) FOXC1 and (g) ERBB2. Since very few genes associated with cell division and proliferation are part of the 'intrinsic' gene list, such a cluster was selected from the total list of genes (Additional file 4), clustered and organized according to (h) the 'intrinsic' dendrogram to show the difference in proliferation between the two luminal groups. (i) In the same manner, a gene cluster characteristic of the mucinous breast carcinomas was made from the total list of genes.
Figure 2
Correlation with five centroids. The expression profiles of the samples were correlated with five previously defined centroids (listed with color codes). The correlation coefficients were plotted according to the dendrogram in Figure 1 and a continuous and opposite curve-pattern of luminal A versus basal-like is evident. The subcluster of luminal samples with the second highest correlation with the luminal B centroid is named 'highly proliferating luminals' in our study.
Figure 3
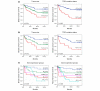
Kaplan-Meier survival curves. (a) Kaplan-Meier plots of breast cancer survival for all patients. The left panel shows tumor size (T1, T2, T3+T4) and the right panel TP53 mutation status (WT, wild type; MUT, mutation). (b) Kaplan-Meier plots of breast cancer survival for patients with gene expression data; the left panel shows tumor size and the right panel TP53 mutation status. (c) Kaplan-Meier plots of breast cancer survival and recurrence-free survival according to gene expression group (LA, luminal A; NL, normal-like; ERBB2; BL, basal-like; HPL, highly proliferating luminals). The p value from the log rank test (Mantel-Cox) is shown in each panel; 'n' is the number of samples in each group. Deaths due to causes not related to breast cancer were treated as censored observations.
Figure 4
Clinical, histopathological and molecular characteristics of subgroups. Dendrogram from hierarchical clustering with distribution of clinical, histopathological and molecular markers between the five gene expression groups (highly proliferating luminals, luminal A, normal-like, basal-like, and ERBB2+). The color coding is as follows: red, description to the left; gray, unknown; yellow, all other. P values from cross-tabulation and Pearson _X_2-test are shown to the right of the panel. Relative expression of mRNA is shown for TP53 (Clone-ID: IMAGE:24415 and IMAGE:236338) and estrogen receptor (ER) (IMAGE:725321). The fraction of malignant cells in tumor tissue and histological type are shown in the lower panel: DCIS, ductal carcinoma in situ with microinvasion IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; MPC, metaplastic carcinoma; MUC, mucinous carcinoma; TLC, tubulolobular carcinoma. ILC* is the 'typical lobular' type previously described [36]. IHC, immunohistochemistry; LOH, loss of heterozygosity.
Figure 5
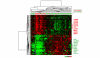
Gene expression pattern associated with TP53 mutations status. Hierarchical clustering of 80 samples using 80 genes selected by significance analysis of microarray analysis to be associated with TP53 mutation status. Tumor samples with TP53 mutation are labeled red and wild-type samples are labeled green (upper dendrogram). Ten genes with the highest correlation in each of the two main branches of the gene cluster (left dendrogram) are listed on the right.
Similar articles
- Combined Immunohistochemistry of PLK1, p21, and p53 for Predicting TP53 Status: An Independent Prognostic Factor of Breast Cancer.
Watanabe G, Ishida T, Furuta A, Takahashi S, Watanabe M, Nakata H, Kato S, Ishioka C, Ohuchi N. Watanabe G, et al. Am J Surg Pathol. 2015 Aug;39(8):1026-34. doi: 10.1097/PAS.0000000000000386. Am J Surg Pathol. 2015. PMID: 26171916 - TP53 mutation is an independent prognostic marker for poor outcome in both node-negative and node-positive breast cancer.
Overgaard J, Yilmaz M, Guldberg P, Hansen LL, Alsner J. Overgaard J, et al. Acta Oncol. 2000;39(3):327-33. doi: 10.1080/028418600750013096. Acta Oncol. 2000. PMID: 10987229 - Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen.
Bertheau P, Turpin E, Rickman DS, Espié M, de Reyniès A, Feugeas JP, Plassa LF, Soliman H, Varna M, de Roquancourt A, Lehmann-Che J, Beuzard Y, Marty M, Misset JL, Janin A, de Thé H. Bertheau P, et al. PLoS Med. 2007 Mar;4(3):e90. doi: 10.1371/journal.pmed.0040090. PLoS Med. 2007. PMID: 17388661 Free PMC article. - p53 in breast cancer subtypes and new insights into response to chemotherapy.
Bertheau P, Lehmann-Che J, Varna M, Dumay A, Poirot B, Porcher R, Turpin E, Plassa LF, de Roquancourt A, Bourstyn E, de Cremoux P, Janin A, Giacchetti S, Espié M, de Thé H. Bertheau P, et al. Breast. 2013 Aug;22 Suppl 2:S27-9. doi: 10.1016/j.breast.2013.07.005. Breast. 2013. PMID: 24074787 Review. - Overview on human breast cancer with focus on prognostic and predictive factors with special attention on the tumour suppressor gene p53.
Norberg T, Jansson T, Sjøgren S, Mårtensson C, Andréasson I, Fjällskog ML, Lindman H, Nordgren H, Lindgren A, Holmberg L, Bergh J. Norberg T, et al. Acta Oncol. 1996;35 Suppl 5:96-102. doi: 10.3109/02841869609083980. Acta Oncol. 1996. PMID: 9142977 Review.
Cited by
- Invasive Breast Cancer: Recognition of Molecular Subtypes.
Strehl JD, Wachter DL, Fasching PA, Beckmann MW, Hartmann A. Strehl JD, et al. Breast Care (Basel). 2011;6(4):258-264. doi: 10.1159/000331339. Epub 2011 Aug 26. Breast Care (Basel). 2011. PMID: 22135623 Free PMC article. - Minireview: Basal-like breast cancer: from molecular profiles to targeted therapies.
Toft DJ, Cryns VL. Toft DJ, et al. Mol Endocrinol. 2011 Feb;25(2):199-211. doi: 10.1210/me.2010-0164. Epub 2010 Sep 22. Mol Endocrinol. 2011. PMID: 20861225 Free PMC article. Review. - TP53 mutations in human cancers: origins, consequences, and clinical use.
Olivier M, Hollstein M, Hainaut P. Olivier M, et al. Cold Spring Harb Perspect Biol. 2010 Jan;2(1):a001008. doi: 10.1101/cshperspect.a001008. Cold Spring Harb Perspect Biol. 2010. PMID: 20182602 Free PMC article. Review. - Early onset HER2-positive breast cancer is associated with germline TP53 mutations.
Melhem-Bertrandt A, Bojadzieva J, Ready KJ, Obeid E, Liu DD, Gutierrez-Barrera AM, Litton JK, Olopade OI, Hortobagyi GN, Strong LC, Arun BK. Melhem-Bertrandt A, et al. Cancer. 2012 Feb 15;118(4):908-13. doi: 10.1002/cncr.26377. Epub 2011 Jul 14. Cancer. 2012. PMID: 21761402 Free PMC article. - TP53 mutations and SNPs as prognostic and predictive factors in patients with breast cancer.
Huszno J, Grzybowska E. Huszno J, et al. Oncol Lett. 2018 Jul;16(1):34-40. doi: 10.3892/ol.2018.8627. Epub 2018 May 3. Oncol Lett. 2018. PMID: 29928384 Free PMC article. Review.
References
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de RM, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous