A semi-quantitative GeLC-MS analysis of temporal proteome expression in the emerging nosocomial pathogen Ochrobactrum anthropi - PubMed (original) (raw)
A semi-quantitative GeLC-MS analysis of temporal proteome expression in the emerging nosocomial pathogen Ochrobactrum anthropi
Robert Leslie James Graham et al. Genome Biol. 2007.
Abstract
Background: The alpha-Proteobacteria are capable of interaction with eukaryotic cells, with some members, such as Ochrobactrum anthropi, capable of acting as human pathogens. O. anthropi has been the cause of a growing number of hospital-acquired infections; however, little is known about its growth, physiology and metabolism. We used proteomics to investigate how protein expression of this organism changes with time during growth.
Results: This first gel-based liquid chromatography-mass spectrometry (GeLC-MS) temporal proteomic analysis of O. anthropi led to the positive identification of 131 proteins. These were functionally classified and physiochemically characterized. Utilizing the emPAI protocol to estimate protein abundance, we assigned molar concentrations to all proteins, and thus were able to identify 19 with significant changes in their expression. Pathway reconstruction led to the identification of a variety of central metabolic pathways, including nucleotide biosynthesis, fatty acid anabolism, glycolysis, TCA cycle and amino acid metabolism. In late phase growth we identified a number of gene products under the control of the oxyR regulon, which is induced in response to oxidative stress and whose protein products have been linked with pathogen survival in response to host immunity reactions.
Conclusion: This study identified distinct proteomic profiles associated with specific growth points for O. anthropi, while the use of emPAI allowed semi-quantitative analyses of protein expression. It was possible to reconstruct central metabolic pathways and infer unique functional and adaptive processes associated with specific growth phases, thereby resulting in a deeper understanding of the physiology and metabolism of this emerging pathogenic bacterium.
Figures
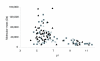
Figure 1
Theoretical two-dimensional map of the soluble sub-proteome of O. anthropi. Diamonds, early growth phase; squares, both growth conditions; triangles, late growth phase.
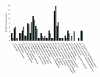
Figure 2
Functional categorisation of identified proteins from the soluble sub-proteome of O. anthropi. Gray bars, early growth phase; black bars, late growth phase.
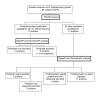
Figure 3
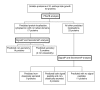
Overview of identified proteins from the soluble sub-proteome of O. anthropi at the early growth phase. Cellular localization was predicted based upon the use of PSortB v2.0.4 [41,42], SignalP v3.0 [43,44], and SecretomeP v2.0 [45,46].
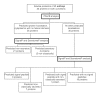
Figure 4
Overview of identified proteins from the soluble sub-proteome of O. anthropi present in both growth conditions. Cellular localization was predicted based upon the use of PSortB v2.0.4 [41,42], SignalP v3.0 [43,44], and SecretomeP v2.0 [45,46].
Figure 5
Overview of identified proteins from the soluble sub-proteome of O. anthropi present in late growth conditions. Cellular localization was predicted based upon the use of PSortB v2.0.4 [41,42], SignalP v3.0 [43,44], and SecretomeP v2.0 [45,46].
Figure 6
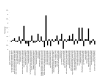
Differential expression profile of proteins common to both growth phases of O. anthropi. Fold changes of ≥1.5 or ≤0.5 are significant.
Figure 7
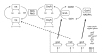
Proposed model for induction of the OxyR regulon in O. anthropi. Oxidized oxyR regulates expression of the OxyR regulon in response to oxidative and nitrosative stress, inducing trxC, grxA, gorA and other OxyR regulon genes. ahpC, alkyl hydroperodide reductase I; fhuF, ferric reductase; fur, ferric uptake repressor; GSSH/GSH, oxidized/reduced glutathione; GorA, glutathione reductase; GrxA, glutaredoxin A; katG, catalase (hydroperoxidase I); OMP agn43, outer membrane protein; oxyS, regulatory RNA; RNO, reactive nitrogen species; ROS, reactive oxygen species; TP, thiol peroxidase; TR, thioredoxin 2. (Adapted (with kind permission of Springer Science and Business Media) from Figure 2a [70].)
Similar articles
- Multidimensional proteomic analysis of the soluble subproteome of the emerging nosocomial pathogen Ochrobactrum anthropi.
Graham RL, Pollock CE, O'Loughlin SN, Ternan NG, Weatherly DB, Jackson PJ, Tarleton RL, McMullan G. Graham RL, et al. J Proteome Res. 2006 Nov;5(11):3145-53. doi: 10.1021/pr060293g. J Proteome Res. 2006. PMID: 17081066 - Temperature-dependent regulation of the Ochrobactrum anthropi proteome.
Varano M, Gaspari M, Quirino A, Cuda G, Liberto MC, Focà A. Varano M, et al. Proteomics. 2016 Dec;16(23):3019-3024. doi: 10.1002/pmic.201600048. Proteomics. 2016. PMID: 27753207 - Cross-species characterisation of abundantly expressed Ochrobactrum anthropi gene products.
Wasinger VC, Urquhart BL, Humphery-Smith I. Wasinger VC, et al. Electrophoresis. 1999 Aug;20(11):2196-203. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2196::AID-ELPS2196>3.0.CO;2-V. Electrophoresis. 1999. PMID: 10493124 - [Research progress of Ochrobactrum anthropic--A review].
Liu Z, Cui B, Xia X. Liu Z, et al. Wei Sheng Wu Xue Bao. 2015 Aug 4;55(8):977-82. Wei Sheng Wu Xue Bao. 2015. PMID: 26665594 Review. Chinese. - Prosthetic mitral valve endocarditis due to Ochrobactrum anthropi: case report.
Romero Gómez MP, Peinado Esteban AM, Sobrino Daza JA, Sáez Nieto JA, Alvarez D, Peña García P. Romero Gómez MP, et al. J Clin Microbiol. 2004 Jul;42(7):3371-3. doi: 10.1128/JCM.42.7.3371-3373.2004. J Clin Microbiol. 2004. PMID: 15243121 Free PMC article. Review.
Cited by
- Curcuminoid supplementation in canine diabetic mellitus and its complications using proteomic analysis.
Suemanotham N, Photcharatinnakorn P, Chantong B, Buranasinsup S, Phochantachinda S, Sakcamduang W, Reamtong O, Thiangtrongjit T, Chatchaisak D. Suemanotham N, et al. Front Vet Sci. 2022 Dec 23;9:1057972. doi: 10.3389/fvets.2022.1057972. eCollection 2022. Front Vet Sci. 2022. PMID: 36619946 Free PMC article. - Colonization by endophytic Ochrobactrum anthropi Mn1 promotes growth of Jerusalem artichoke.
Meng X, Yan D, Long X, Wang C, Liu Z, Rengel Z. Meng X, et al. Microb Biotechnol. 2014 Nov;7(6):601-10. doi: 10.1111/1751-7915.12145. Epub 2014 Jul 30. Microb Biotechnol. 2014. PMID: 25073416 Free PMC article. - Semiquantitative analysis of clinical heat stress in Clostridium difficile strain 630 using a GeLC/MS workflow with emPAI quantitation.
Ternan NG, Jain S, Graham RL, McMullan G. Ternan NG, et al. PLoS One. 2014 Feb 24;9(2):e88960. doi: 10.1371/journal.pone.0088960. eCollection 2014. PLoS One. 2014. PMID: 24586458 Free PMC article. - Comparative proteome analysis of Milnesium tardigradum in early embryonic state versus adults in active and anhydrobiotic state.
Schokraie E, Warnken U, Hotz-Wagenblatt A, Grohme MA, Hengherr S, Förster F, Schill RO, Frohme M, Dandekar T, Schnölzer M. Schokraie E, et al. PLoS One. 2012;7(9):e45682. doi: 10.1371/journal.pone.0045682. Epub 2012 Sep 27. PLoS One. 2012. PMID: 23029181 Free PMC article. - An economical high-throughput protocol for multidimensional fractionation of proteins.
Tooth DJ, Gopala Krishna V, Layfield R. Tooth DJ, et al. Int J Proteomics. 2012;2012:735132. doi: 10.1155/2012/735132. Epub 2012 Sep 12. Int J Proteomics. 2012. PMID: 23008771 Free PMC article.
References
- Teyssier C, Marchandin H, Simeon De Buochberg M, Ramuz M, Jumas-Bilak E. Atypical 16S rRNA gene copies in Ochrobactrum intermedium strains reveal a large genomic rearrangement by recombination between rrn copies. J Bacteriol. 2003;185:2901–2909. doi: 10.1128/JB.185.9.2901-2909.2003. - DOI - PMC - PubMed
- Holmes B, Popoff M, Kiredjian M, Kersters K. Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as group vd. Int J Syst Bacteriol. 1988;38:406–416.
- Velasco J, Romero C, Lopez-Goni I, Leiva J, Diaz R, Moriyon I. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int J Syst Bacteriol. 1998;48:759–768. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources