Detailed analysis of 15q11-q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader-Willi, Angelman, and inv dup(15) syndromes - PubMed (original) (raw)
Detailed analysis of 15q11-q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader-Willi, Angelman, and inv dup(15) syndromes
Andrew J Makoff et al. Genome Biol. 2007.
Abstract
Background: Chromosome 15 contains many segmental duplications, including some at 15q11-q13 that appear to be responsible for the deletions that cause Prader-Willi and Angelman syndromes and for other genomic disorders. The current version of the human genome sequence is incomplete, with seven gaps in the proximal region of 15q, some of which are flanked by duplicated sequence. We have investigated this region by conducting a detailed examination of the sequenced genomic clones in the public database, focusing on clones from the RP11 library that originates from one individual.
Results: Our analysis has revealed assembly errors, including contig NT_078094 being in the wrong orientation, and has enabled most of the gaps between contigs to be closed. We have constructed a map in which segmental duplications are no longer interrupted by gaps and which together reveals a complex region. There are two pairs of large direct repeats that are located in regions consistent with the two classes of deletions associated with Prader-Willi and Angelman syndromes. There are also large inverted repeats that account for the formation of the observed supernumerary marker chromosomes containing two copies of the proximal end of 15q and associated with autism spectrum disorders when involving duplications of maternal origin (inv dup[15] syndrome).
Conclusion: We have produced a segmental map of 15q11-q14 that reveals several large direct and inverted repeats that are incompletely and inaccurately represented on the current human genome sequence. Some of these repeats are clearly responsible for deletions and duplications in known genomic disorders, whereas some may increase susceptibility to other disorders.
Figures
Figure 1
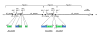
Map showing an overview of build 36 for 15q11-q14. The positions and orientations of the proximal eight contigs of 15q are shown as in build 36, with the HERC2 duplications (segments P, V, and Y) shown in detail. The asterisk above segment V of RP11-536P16 is to indicate that its orientation is shown as in the database. The positions of the seven gaps are shown with the approximate positions of the PWS/AS breakpoint (BP)1 to BP3. The map is divided into three parts for analysis in Figures 2, 3 and 5, as indicated. Mb, megabases.
Figure 2
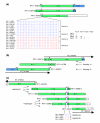
Map of 15q13-q14 at proximal end of contig NT_010194. This part of the map is an updated version of the same region that we analyzed previously [20], with some differences in segment labeling. RP11 clones representing the two possible haplotypes are arbitrarily placed either above or immediately below the segments, with the non-RP11 clones placed below the contig label. Asterisks indicate representative clones used in the contig. Solid lines indicate completely sequenced clones, and dotted lines indicate draft sequences (high throughput genomic sequences [htgs]). A solid line with a dotted line extension indicates a clone in which only a part has been completely sequenced. A gap in a clone indicates a deletion. kb, kilobases.
Figure 3
Map of contigs NT_078095, NT_010280, and NT_078096 (15q12-q13). The clones are indicated as in Figure 2. kb, kilobases.
Figure 5
Map of contigs NT_037852, NT_077631, NT_078094, and part of NT_026446 (15q11-q12). The clones are indicated as in Figure 2. The shaded segment indicates α-satellite DNA sequence. Note that clones CTD-2298I13, CTC-803A3, and 386A2 occur twice to indicate two possible locations with respect to the RP11 sequence. kb, kilobases.
Figure 4
Alignment of 15q11-q13 clones in duplicons adjacent to segment V. (a) The three representative clones containing segment V are aligned, with single nucleotide variants in a 3,356 base pair (bp) region of segment V in all sequenced RP11 clones shown below. The asterisk above segment V indicates its orientation, as in Figure 1. The box shows the number of mismatches between each pair of haplotigs. (b) Corrected alignment of clones to show true relationship between ends of contigs NT_010280 and NT_078096. The hash above segment V of RP11-536P16 is to indicate that its orientation has been inverted compared with that in the database. (c) Alignment of clones around the segment V end of contig NT_078094, with single nucleotide variants in a 9.5 kilobase (kb) region around the small segment P shown below.
Figure 6
Analysis of symmetrical region near the centromeric end of 15q to identify its likeliest arrangement in RP11. The region between the most proximal segments P ordered as in Figure 5 is indicated by the four rows of segments at the top. The first row, continuing to the third row, represents the upper RP11 haplotigs in Figure 5 and the second row, continuing to the fourth row, represents the lower haplotigs. The RP11 haplotigs are shown below the segments with the non-RP11 clones shown further below. Nine slices of 5 to 30 kilobases (kb), shown by alternating red or blue lines, were investigated, with each box showing the number of single nucleotide mismatches between each pair of RP11 haplotigs and non-RP11 clones in the slice.
Figure 7
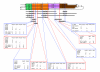
Map showing positions of segmental duplications of 15q11-14 in the RP11 individual. The main part of the map shows the segmental duplications as in Figures 2, 3 and 5, with the approximate positions of genes, duplications (dup), and pseudogenes (ps) shown underneath and the positions of the three remaining contigs at the bottom. Note that most of the imprinted region in the Prader-Willi/Angelman syndrome critical region is not included. The alternative structure (model B) near the centromere is shown underneath. The duplicated regions in each of the five breakpoint regions (BP1 to BP5) are shown in more detail above the map and include the probable structure for those individuals with a BP2:BP3 inversion. The positions of the major direct and inverted repeats are shown above the detail with arrows in an arbitrary direction.
Similar articles
- Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13).
Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Christian SL, et al. Hum Mol Genet. 1999 Jun;8(6):1025-37. doi: 10.1093/hmg/8.6.1025. Hum Mol Genet. 1999. PMID: 10332034 - Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints.
Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD. Amos-Landgraf JM, et al. Am J Hum Genet. 1999 Aug;65(2):370-86. doi: 10.1086/302510. Am J Hum Genet. 1999. PMID: 10417280 Free PMC article. - Screening for genomic rearrangements and methylation abnormalities of the 15q11-q13 region in autism spectrum disorders.
Depienne C, Moreno-De-Luca D, Heron D, Bouteiller D, Gennetier A, Delorme R, Chaste P, Siffroi JP, Chantot-Bastaraud S, Benyahia B, Trouillard O, Nygren G, Kopp S, Johansson M, Rastam M, Burglen L, Leguern E, Verloes A, Leboyer M, Brice A, Gillberg C, Betancur C. Depienne C, et al. Biol Psychiatry. 2009 Aug 15;66(4):349-59. doi: 10.1016/j.biopsych.2009.01.025. Epub 2009 Mar 17. Biol Psychiatry. 2009. PMID: 19278672 - Prader-Willi, Angelman, and 15q11-q13 Duplication Syndromes.
Kalsner L, Chamberlain SJ. Kalsner L, et al. Pediatr Clin North Am. 2015 Jun;62(3):587-606. doi: 10.1016/j.pcl.2015.03.004. Epub 2015 Apr 22. Pediatr Clin North Am. 2015. PMID: 26022164 Free PMC article. Review. - Genomic imprinting and uniparental disomy in Angelman and Prader-Willi syndromes: a review.
Nicholls RD. Nicholls RD. Am J Med Genet. 1993 Apr 1;46(1):16-25. doi: 10.1002/ajmg.1320460106. Am J Med Genet. 1993. PMID: 8388169 Review.
Cited by
- Genetically complex epilepsies, copy number variants and syndrome constellations.
Mefford HC, Mulley JC. Mefford HC, et al. Genome Med. 2010 Oct 5;2(10):71. doi: 10.1186/gm192. Genome Med. 2010. PMID: 20923578 Free PMC article. - Behavioral characterization of dup15q syndrome: Toward meaningful endpoints for clinical trials.
DiStefano C, Wilson RB, Hyde C, Cook EH, Thibert RL, Reiter LT, Vogel-Farley V, Hipp J, Jeste S. DiStefano C, et al. Am J Med Genet A. 2020 Jan;182(1):71-84. doi: 10.1002/ajmg.a.61385. Epub 2019 Oct 26. Am J Med Genet A. 2020. PMID: 31654560 Free PMC article. - Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome.
Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, Rosenfeld JA, Lamb AN, Sahoo T. Duker AL, et al. Eur J Hum Genet. 2010 Nov;18(11):1196-201. doi: 10.1038/ejhg.2010.102. Epub 2010 Jun 30. Eur J Hum Genet. 2010. PMID: 20588305 Free PMC article. - Multi-ancestry GWAS reveals loci linked to human variation in LINE-1- and Alu-insertion numbers.
Bravo JI, Zhang L, Benayoun BA. Bravo JI, et al. Transl Med Aging. 2025;9:25-40. doi: 10.1016/j.tma.2025.02.001. Epub 2025 Feb 13. Transl Med Aging. 2025. PMID: 40051556 Free PMC article. - Genomic profile of copy number variants on the short arm of human chromosome 8.
Yu S, Fiedler S, Stegner A, Graf WD. Yu S, et al. Eur J Hum Genet. 2010 Oct;18(10):1114-20. doi: 10.1038/ejhg.2010.66. Epub 2010 May 12. Eur J Hum Genet. 2010. PMID: 20461109 Free PMC article.
References
- Wandstrat AE, Schwartz S. Isolation and molecular analysis of inv dup(15) and construction of a physical map of a common breakpoint in order to elucidate their mechanism of formation. Chromosoma. 2000;109:498–505. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials