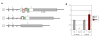Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets - PubMed (original) (raw)
Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets
ChaRandle Jordan et al. BMC Med Genet. 2007.
Abstract
Background: MeCP2, methyl-CpG-binding protein 2, binds to methylated cytosines at CpG dinucleotides, as well as to unmethylated DNA, and affects chromatin condensation. MECP2 mutations in females lead to Rett syndrome, a neurological disorder characterized by developmental stagnation and regression, loss of purposeful hand movements and speech, stereotypic hand movements, deceleration of brain growth, autonomic dysfunction and seizures. Most mutations occur de novo during spermatogenesis. Located at Xq28, MECP2 is subject to X inactivation, and affected females are mosaic. Rare hemizygous males suffer from a severe congenital encephalopathy.
Methods: To identify the pathways mis-regulated by MeCP2 deficiency, microarray-based global gene expression studies were carried out in cerebellum of Mecp2 mutant mice. We compared transcript levels in mutant/wildtype male sibs of two different MeCP2-deficient mouse models at 2, 4 and 8 weeks of age. Increased transcript levels were evaluated by real-time quantitative RT-PCR. Chromatin immunoprecipitation assays were used to document in vivo MeCP2 binding to promoter regions of candidate target genes.
Results: Of several hundred genes with altered expression levels in the mutants, twice as many were increased than decreased, and only 27 were differentially expressed at more than one time point. The number of misregulated genes was 30% lower in mice with the exon 3 deletion (Mecp2tm1.1Jae) than in mice with the larger deletion (Mecp2tm1.1Bird). Between the mutants, few genes overlapped at each time point. Real-time quantitative RT-PCR assays validated increased transcript levels for four genes: Irak1, interleukin-1 receptor-associated kinase 1; Fxyd1, phospholemman, associated with Na, K-ATPase;Reln, encoding an extracellular signaling molecule essential for neuronal lamination and synaptic plasticity; and Gtl2/Meg3, an imprinted maternally expressed non-translated RNA that serves as a host gene for C/D box snoRNAs and microRNAs. Chromatin immunoprecipitation assays documented in vivo MeCP2 binding to promoter regions of Fxyd1, Reln, and Gtl2.
Conclusion: Transcriptional profiling of cerebellum failed to detect significant global changes in Mecp2-mutant mice. Increased transcript levels of Irak1, Fxyd1, Reln, and Gtl2 may contribute to the neuronal dysfunction in MeCP2-deficient mice and individuals with Rett syndrome. Our data provide testable hypotheses for future studies of the regulatory or signaling pathways that these genes act on.
Figures
Figure 1
Structure and expression of Mecp2 mutant alleles. A. Genomic structure of Mecp2 mutations. a. Wild-type Mecp2 gene. MBD, methyl-CpG binding domain; TRD, transcription repression domain. White or hatched: coding sequence. Grey: non-coding parts of transcript. Exons 1 and 2 that are alternatively spliced, and intron 2, are not drawn to scale. Horizontal bar above exon 4 denotes location of the amplicon analyzed by qRT-PCR (in Figure 1B) b. The _Mecp2_tm1.1Jae (J-allele) mutant has a deletion of exon 3 that encodes the N-terminal half of the MBD. c. In the _Mecp2_tm1.1Bird (B-allele) mutant, exon 3, the coding portion of exon 4 and part of the 3'UTR are deleted. This larger deletion eliminates ~ 97% of the coding sequence. B. Real-time quantitative RT-PCR analysis of Mecp2 expression in cerebellum. Using primers within the deleted region, Mecp2 transcript was undetectable in mutant mice with the B-allele, when four wild-type and four mutant samples were compared at 8 wk. This proves the specificity of the primers. In mice with the J-allele, mean Mecp2 expression was higher in the mutants, but this difference was not statistically significant (P = 0.11), when six wild-type and seven mutant samples were compared at 8 wk.
Figure 2
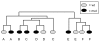
Hierarchical clustering of microarray data from six mutant/wildtype sib pairs. Male B-allele mutants at 8 wk of age, when most of them are symptomatic, tended to cluster with their wt sibs rather than with mutants of other litters. Litters are identified by A – F. The array of the litter B wild-type sib did not pass quality control standards.
Figure 3
Hierarchical cluster analysis of microarray data from wild-type sibs at three different ages. Strain background had the greatest influence on clustering. BALB/c is the major background of the J-allele mice, while B-allele mice are on a C57BL/6 background. The effects of array batch, and to a lesser extent, age, are also apparent. We, therefore, compared only microarrays from littermates, at the same age and using the same array batch, in our search for differentially expressed genes.
Figure 4
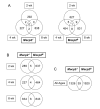
Differentially expressed genes (DEG) in cerebellum microarray study. A. Overlap of DEG at 2 wk, 4 wk, and 8 wk time points. For both mutant alleles, genes whose transcripts were either increased (1.2-fold or higher) or decreased (0.8-fold or lower) at P < 0.05 are included. Transcripts in the overlap regions are described in Additional file 1. B. Overlap of DEG across both mutant alleles at three time points. The same criteria as in Figure 4A were applied. C. Overlap of DEG across all ages in both mutant strains. The 39 transcripts include the 15 overlapping DEG from Figure 4B as well as genes that were significantly changed at one age in one mutant and at another time point in the other mutant. The list is provided in the Additional file 1.
Figure 5
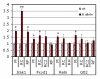
Microarray and qRT-PCR results for select DEGs. Irak1, Fxyd1, Reln and Gtl2 were selected for validation of expression changes by qRT-PCR analyses in B-allele mice. Cerebellum microarray expression data (M) are compared with qRT-PCR data from cerebellum (RC), forebrain (RF) and hippocampus (RH). For RC of Irak1, Fxyd1 and Gtl2, the 8 wk samples from the microarray analysis were used. RC2 data are derived from an independent set of litters at 7.5 to 8 wk. Error bars represent SEM. One asterisk indicates P < 0.05, two asterisks indicate P < 0.001. Irak1 expression was markedly increased in the cerebellum of 8 wk B-allele mice on microarray (2-fold, P = 0.011, four mutant/wild-type pairs) and qRT-PCR (3.5 fold, P < 0.001, four mutant/wild-type pairs) as well as in the forebrain of a different set of mice at 7.5 wk (1.9 fold, P < 0.05, three mutant/five wild-type). Fxyd1 expression was elevated 1.4-fold on microarrays (P < 0.05, five mutant/wild-type sibs) and qRT-PCR (RC) (P = 0.053, four mutant/wild-type sibs). The RC2 data (five mutant/three wild-type) were more variable and yielded P = 0.16. Reln displayed an ~ 1.5 fold increase in expression on microarrays (P = 0.03, four mutant/wild-type pairs) and qRT-PCR (P = 0.06, five mutant/wild-type pairs) at 2 wk (RC) and was not changed in hippocampus (RH). On microarray, Gtl2 expression was increased at 8 wk, but by qRT-PCR we found highly variable Gtl2 expression in cerebellum (RC and RC2). Gtl2 expression was not significantly increased (P = 0.29, eight mutant/eight wild-type) in the forebrain.
Figure 6
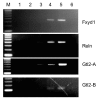
Chromatin immunoprecipitation (ChIP) analysis of differentially expressed genes. Whole brain nuclear extracts from 4 wk old wild-type and B-allele mutants were used for ChIP. Lane M, 100 bp ladder; lane 1, _Mecp2_-mutant chromatin precipitated with anti-MeCP2 (negative control); lane 2, wild-type chromatin precipitated with no antibody present; lane 3, wild-type chromatin precipitated with rabbit IgG, lane 4: wild-type chromatin precipitated with anti-MeCP2, lane 5: input DNA, lane 6: no DNA control. The precipitated DNA was amplified with primers specific for the promoter regions of Fxyd1 and Reln, and for the promoter/DMR of Gtl2. The Gtl2-B amplicon is located within the Gtl2-A amplicon. Precipitation with anti-MeCP2 shows enrichment relative to the IgG and no antibody controls, indicating that MeCP2 is associated with the designated regions in vivo. As negative controls, PCR was performed using primers covering regions that do not interact with MeCP2 for each ChIP reaction (not shown).
Figure 7
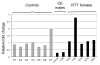
Relative FXYD1 expression in human frontal cortex. Quantitative real-time RT-PCR analyses were carried out on frozen frontal cortex samples from seven unaffected controls (C1 – C7), two _MECP2_-mutant males with congenital encephalopathy (CE) and five females diagnosed with RTT, with or without identified MECP2 mutations. The mutant samples are identified by their brain bank numbers. FXYD1 expression was normalized to RPS18, a constitutive ribosomal protein gene that is expressed at a similar level in brain. To calculate relative fold changes, control sample C4 was arbitrarily set at 1. All samples were run in triplicate on the same plate.
Similar articles
- FXYD1 is an MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice.
Deng V, Matagne V, Banine F, Frerking M, Ohliger P, Budden S, Pevsner J, Dissen GA, Sherman LS, Ojeda SR. Deng V, et al. Hum Mol Genet. 2007 Mar 15;16(6):640-50. doi: 10.1093/hmg/ddm007. Epub 2007 Feb 19. Hum Mol Genet. 2007. PMID: 17309881 - Astrocyte Transcriptome from the Mecp2(308)-Truncated Mouse Model of Rett Syndrome.
Delépine C, Nectoux J, Letourneur F, Baud V, Chelly J, Billuart P, Bienvenu T. Delépine C, et al. Neuromolecular Med. 2015 Dec;17(4):353-63. doi: 10.1007/s12017-015-8363-9. Epub 2015 Jul 25. Neuromolecular Med. 2015. PMID: 26208914 - Ube3a expression is not altered in Mecp2 mutant mice.
Jordan C, Francke U. Jordan C, et al. Hum Mol Genet. 2006 Jul 15;15(14):2210-5. doi: 10.1093/hmg/ddl146. Epub 2006 Jun 5. Hum Mol Genet. 2006. PMID: 16754645 - MeCP2 dysfunction in Rett syndrome and related disorders.
Moretti P, Zoghbi HY. Moretti P, et al. Curr Opin Genet Dev. 2006 Jun;16(3):276-81. doi: 10.1016/j.gde.2006.04.009. Epub 2006 May 2. Curr Opin Genet Dev. 2006. PMID: 16647848 Review. - Associations between MeCP2 mutations, X-chromosome inactivation, and phenotype.
Hoffbuhr KC, Moses LM, Jerdonek MA, Naidu S, Hoffman EP. Hoffbuhr KC, et al. Ment Retard Dev Disabil Res Rev. 2002;8(2):99-105. doi: 10.1002/mrdd.10026. Ment Retard Dev Disabil Res Rev. 2002. PMID: 12112735 Review.
Cited by
- Genetic modifiers of MeCP2 function in Drosophila.
Cukier HN, Perez AM, Collins AL, Zhou Z, Zoghbi HY, Botas J. Cukier HN, et al. PLoS Genet. 2008 Sep 5;4(9):e1000179. doi: 10.1371/journal.pgen.1000179. PLoS Genet. 2008. PMID: 18773074 Free PMC article. - Accelerated Hyper-Maturation of Parvalbumin Circuits in the Absence of MeCP2.
Patrizi A, Awad PN, Chattopadhyaya B, Li C, Di Cristo G, Fagiolini M. Patrizi A, et al. Cereb Cortex. 2020 Jan 10;30(1):256-268. doi: 10.1093/cercor/bhz085. Cereb Cortex. 2020. PMID: 31038696 Free PMC article. - Microglia as a critical player in both developmental and late-life CNS pathologies.
Derecki NC, Katzmarski N, Kipnis J, Meyer-Luehmann M. Derecki NC, et al. Acta Neuropathol. 2014 Sep;128(3):333-45. doi: 10.1007/s00401-014-1321-z. Epub 2014 Jul 24. Acta Neuropathol. 2014. PMID: 25056803 Free PMC article. Review. - Rett syndrome: a neurological disorder with metabolic components.
Kyle SM, Vashi N, Justice MJ. Kyle SM, et al. Open Biol. 2018 Feb;8(2):170216. doi: 10.1098/rsob.170216. Open Biol. 2018. PMID: 29445033 Free PMC article. Review. - MeCP2 mutation results in compartment-specific reductions in dendritic branching and spine density in layer 5 motor cortical neurons of YFP-H mice.
Stuss DP, Boyd JD, Levin DB, Delaney KR. Stuss DP, et al. PLoS One. 2012;7(3):e31896. doi: 10.1371/journal.pone.0031896. Epub 2012 Mar 7. PLoS One. 2012. PMID: 22412847 Free PMC article.
References
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases