Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample - PubMed (original) (raw)
Comparative Study
Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample
Shuji Ogino et al. J Mol Diagn. 2007 Jul.
Abstract
The CpG island methylator phenotype (CIMP or CIMP-high) with extensive promoter methylation is a distinct phenotype in colorectal cancer. However, a choice of markers for CIMP has been controversial. A recent extensive investigation has selected five methylation markers (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1) as surrogate markers for epigenomic aberrations in tumor. The use of these markers as a CIMP-specific panel needs to be validated by an independent, large dataset. Using MethyLight assays on 920 colorectal cancers from two large prospective cohort studies, we quantified DNA methylation in eight CIMP-specific markers [the above five plus CDKN2A (p16), CRABP1, and MLH1]. A CIMP-high cutoff was set at > or = 6/8 or > or = 5/8 methylated promoters, based on tumor distribution and BRAF/KRAS mutation frequencies. All but two very specific markers [MLH1 (98% specific) and SOCS1 (93% specific)] demonstrated > or = 85% sensitivity and > or = 80% specificity, indicating overall good concordance in methylation patterns and good performance of these markers. Based on sensitivity, specificity, and false positives and negatives, the eight markers were ranked in order as: RUNX3, CACNA1G, IGF2, MLH1, NEUROG1, CRABP1, SOCS1, and CDKN2A. In conclusion, a panel of markers including at least RUNX3, CACNA1G, IGF2, and MLH1 can serve as a sensitive and specific marker panel for CIMP-high.
Figures
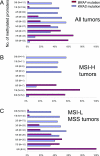
Figure 1
BRAF and KRAS mutation frequencies according to number of methylated promoters. A: Tumors with ≥6/8 methylated promoters show high BRAF mutation rates, whereas tumors with ≤5/8 methylated promoters show high KRAS mutation rates. B: MSI-H tumors distribute bimodally, and the frequencies of KRAS and BRAF mutations clearly distinguish CIMP-high tumors from CIMP-low tumors. C: MSI-L/MSS tumors can be separated into CIMP-high (≥6/8 methylated promoters) and CIMP-low/0 (≤4/8 methylated promoters) based on the frequencies of BRAF and KRAS mutations. Tumors with 5/8 methylated promoters reside on the borderline between CIMP-high and CIMP-low.
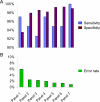
Figure 2
Sensitivity, specificity, and cross-panel classification error rate against panel 8. Panel 1 (RUNX3 only) through panel 7 contain incrementing numbers of markers, adding one by one from CACNA1G, IGF2, MLH1, NEUROG1, CRABP1, and SOCS1. Panel 8 contains all eight markers including CDKN2A. A: Specificity generally increases with an increasing number of markers. Sensitivity depends on the number of markers and a CIMP-high cutoff. B: The classification error rate decreases with an increasing number of markers.
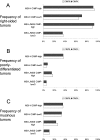
Figure 3
Frequencies of right-sided tumors (A), poorly differentiated tumors (B), and mucinous tumors (C) in various MSI/CIMP subtypes of colorectal cancer. Gray and open bar graphs indicate frequencies of each feature in MSI/CIMP subtypes determined by CIMP panel 4 and CIMP panel 8, respectively. Note that there were no substantial differences in the features examined (anatomical location, tumor grade, or mucinous features) between classifications determined by CIMP panel 4 and panel 8.
Comment in
- The CpG island methylator phenotype in colorectal cancer.
Samowitz WS. Samowitz WS. J Mol Diagn. 2007 Jul;9(3):281-3. doi: 10.2353/jmoldx.2007.070031. J Mol Diagn. 2007. PMID: 17591925 Free PMC article. No abstract available.
Similar articles
- CpG island methylator phenotype-low (CIMP-low) colorectal cancer shows not only few methylated CIMP-high-specific CpG islands, but also low-level methylation at individual loci.
Kawasaki T, Ohnishi M, Nosho K, Suemoto Y, Kirkner GJ, Meyerhardt JA, Fuchs CS, Ogino S. Kawasaki T, et al. Mod Pathol. 2008 Mar;21(3):245-55. doi: 10.1038/modpathol.3800982. Epub 2008 Jan 18. Mod Pathol. 2008. PMID: 18204436 Clinical Trial. - TGFBR2 mutation is correlated with CpG island methylator phenotype in microsatellite instability-high colorectal cancer.
Ogino S, Kawasaki T, Ogawa A, Kirkner GJ, Loda M, Fuchs CS. Ogino S, et al. Hum Pathol. 2007 Apr;38(4):614-20. doi: 10.1016/j.humpath.2006.10.005. Epub 2007 Jan 31. Hum Pathol. 2007. PMID: 17270239 - Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample.
Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D, Giovannucci EL, Fuchs CS, Ogino S. Nosho K, et al. PLoS One. 2008;3(11):e3698. doi: 10.1371/journal.pone.0003698. Epub 2008 Nov 12. PLoS One. 2008. PMID: 19002263 Free PMC article. - Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review.
Shima K, Nosho K, Baba Y, Cantor M, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. Shima K, et al. Int J Cancer. 2011 Mar 1;128(5):1080-94. doi: 10.1002/ijc.25432. Int J Cancer. 2011. PMID: 20473920 Free PMC article. Review. - Global differences in the prevalence of the CpG island methylator phenotype of colorectal cancer.
Advani SM, Advani PS, Brown DW, DeSantis SM, Korphaisarn K, VonVille HM, Bressler J, Lopez DS, Davis JS, Daniel CR, Sarshekeh AM, Braithwaite D, Swartz MD, Kopetz S. Advani SM, et al. BMC Cancer. 2019 Oct 17;19(1):964. doi: 10.1186/s12885-019-6144-9. BMC Cancer. 2019. PMID: 31623592 Free PMC article.
Cited by
- DNA hypermethylation appears early and shows increased frequency with dysplasia in Lynch syndrome-associated colorectal adenomas and carcinomas.
Valo S, Kaur S, Ristimäki A, Renkonen-Sinisalo L, Järvinen H, Mecklin JP, Nyström M, Peltomäki P. Valo S, et al. Clin Epigenetics. 2015 Jul 22;7(1):71. doi: 10.1186/s13148-015-0102-4. eCollection 2015. Clin Epigenetics. 2015. PMID: 26203307 Free PMC article. - Distinct patterns of DNA methylation in conventional adenomas involving the right and left colon.
Koestler DC, Li J, Baron JA, Tsongalis GJ, Butterly LF, Goodrich M, Lesseur C, Karagas MR, Marsit CJ, Moore JH, Andrew AS, Srivastava A. Koestler DC, et al. Mod Pathol. 2014 Jan;27(1):145-55. doi: 10.1038/modpathol.2013.104. Epub 2013 Jul 19. Mod Pathol. 2014. PMID: 23868178 Free PMC article. - CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma.
Ogino S, Meyerhardt JA, Kawasaki T, Clark JW, Ryan DP, Kulke MH, Enzinger PC, Wolpin BM, Loda M, Fuchs CS. Ogino S, et al. Virchows Arch. 2007 May;450(5):529-37. doi: 10.1007/s00428-007-0398-3. Epub 2007 Mar 20. Virchows Arch. 2007. PMID: 17372756 - Defects in MMR Genes as a Seminal Example of Personalized Medicine: From Diagnosis to Therapy.
Dal Buono A, Gaiani F, Poliani L, Correale C, Laghi L. Dal Buono A, et al. J Pers Med. 2021 Dec 8;11(12):1333. doi: 10.3390/jpm11121333. J Pers Med. 2021. PMID: 34945805 Free PMC article. Review. - Global Methylome Scores Correlate with Histological Subtypes of Colorectal Carcinoma and Show Different Associations with Common Clinical and Molecular Features.
Turpín-Sevilla MDC, Pérez-Sanz F, García-Solano J, Sebastián-León P, Trujillo-Santos J, Carbonell P, Estrada E, Tuomisto A, Herruzo I, Fennell LJ, Mäkinen MJ, Rodríguez-Braun E, Whitehall VLJ, Conesa A, Conesa-Zamora P. Turpín-Sevilla MDC, et al. Cancers (Basel). 2021 Oct 14;13(20):5165. doi: 10.3390/cancers13205165. Cancers (Basel). 2021. PMID: 34680315 Free PMC article.
References
- Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14(Spec No 1):R65–R76. - PubMed
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. - PubMed
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous