Identification and characterization of interferon-induced proteins that inhibit alphavirus replication - PubMed (original) (raw)
Identification and characterization of interferon-induced proteins that inhibit alphavirus replication
Yugen Zhang et al. J Virol. 2007 Oct.
Abstract
Alpha/beta interferon (IFN-alpha/beta) produces antiviral effects through upregulation of many interferon-stimulated genes (ISGs) whose protein products are effectors of the antiviral state. Previous data from our laboratory have shown that IFN-alpha/beta can limit Sindbis virus (SB) replication through protein kinase R (PKR)-dependent and PKR-independent mechanisms and that one PKR-independent mechanism inhibits translation of the infecting virus genome (K. D. Ryman et al., J. Virol. 79:1487-1499, 2005). Further, using Affymetrix microarray technology, we identified 44 genes as candidates for PKR/RNase L-independent IFN-induced antiviral activities. In the current studies, we have begun analyzing these gene products for antialphavirus activity using three techniques: (i) overexpression of the protein from SB vectors and assessment of virulence attenuation in mice; (ii) overexpression of the proteins in a stable tetracycline-inducible murine fibroblast culture system and assessment of effects upon SB replication; and (iii) small interfering RNA-mediated knockdown of gene mRNA in fibroblast cultures followed by SB replication assessment as above. Tested proteins included those we hypothesized had potential to affect virus genome translation and included murine ISG20, ISG15, the zinc finger antiviral protein (ZAP), viperin, p56, p54, and p49. Interestingly, the pattern of antiviral activity for some gene products was different between in vitro and in vivo assays. Viperin and ZAP attenuated virulence most profoundly in mice. However, ISG20 and ZAP potently inhibited SB replication in vitro, whereas and viperin, p56, and ISG15 exhibited modest replication inhibition in vitro. In contrast, p54 and p49 had little to no effect in any assay.
Figures
FIG. 1.
Diagrams of the genomes of wild-type Sindbis virus strain TR339 (A) and the 39MCS double subgenomic promoter virus (B) used for in vivo expression of candidate genes.
FIG. 2.
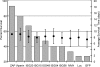
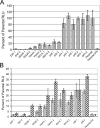
Neonatal mouse mortality and average survival time data from infections with viruses. Mice were inoculated subcutaneously in the axial region at exactly 24 h of age with 5 μg of RNA from an in vitro transcription reaction mixture of each expression virus. Two separate pooled litters totaling 15 to 20 mice were used for each virus. Percent mortality and average survival times were calculated as described in Materials and Methods.
FIG. 3.
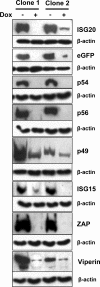
Basal and induced expression of tested proteins in Tet-OFF MEF cultures. Cultures of two separately selected clones expressing each protein were either incubated in doxycycline (+) or incubated without doxycycline (−) for 48 to 72 h followed by lysis, SDS-PAGE separation, and Western blotting for FLAG tag as described in Materials and Methods. Fifty micrograms of protein was loaded per well. Staining for β-actin in the samples confirmed equal loading.
FIG. 4.
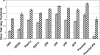
Growth of TR339 at 12 (solid bars) or 24 (hatched bars) hours postinfection (MOI, 0.01) of GFP- or candidate gene-expressing Tet-OFF MEFs or mock-treated (parental) or IFN-α/β-treated, untransfected parental cells (1,000 IU for 6 h). Samples titers were determined by standard plaque assay on BHK cells.
FIG. 5.
RT-PCR for IFN-β mRNA in ISG20- and ZAP-expressing Tet-Off MEF clones 1 and 2 in the presence or absence of DOX and either infected or mock infected with Sendai virus (MOI, 3).
FIG. 6.
Luciferase expression from Tet-OFF MEF cultures at 8 hpi with 39MCS-fLuc. (A) Comparison of all clones with GFP or untransfected (parental) clones or parental cells treated with IFN (1,000 IU/ml for 6 h). (B) Comparison of clones exhibiting an antiviral effect in experiments of panel A either in the presence (hatched bars) or absence (gray bars) of Dox. *, not done.
FIG. 7.
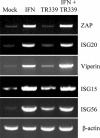
Semiquantitative RT-PCR demonstrating the induction profiles of ZAP, ISG20, viperin, ISG15, and ISG56 mRNAs in Tet-OFF MEF cells after IFN treatment (1,000 IU/ml for 6 h) or TR339 infection (MOI, 3).
FIG. 8.
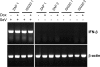
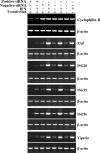
Semiquantitative RT-PCR for mRNAs targeted by siRNA at 48 h post-siRNA transfection. Positive siRNA refers to transfection with the gene-specific RNA listed on the right of the figure in each pair with β-actin. Negative siRNA refers to the presence or absence of a nontargeting control siRNA. In IFN+ samples, cells were treated (at 45 h post-siRNA transfection) with 10 IU of IFN-α/β for 3 h at 37°C followed by sample analysis. “Transfection” refers to the presence or absence of the lipid-based transfection reagent. For each gene, PCR cycles were adjusted such that the maximum signal for a given gene (for example, IFN-treated cells with ZAP, ISG20, ISG15, and p56) was not saturating. Cyclophilin B was included as a control for a non-IFN-inducible mRNA. The β-actin loading control was performed separately for each treatment. PCR primers used for each gene are listed in Table 1.
FIG. 9.
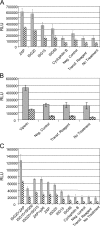
Luciferase expression in Tet-OFF MEF cultures that were mock treated (gray bars) or treated with 10 IU/ml of IFN-α/β for 3 h (hatched bars) prior to infection. Cells were harvested at 8 hpi with 39MCS-fLuc. (A) Comparison of specific siRNA pools targeting ZAP, ISG20, ISG15, and ISG56 with controls, including a cyclophilin B-specific siRNA pool, a nontargeting negative control pool, transfection reagent alone, or no treatment. (B) Comparison of specific siRNA pools targeting viperin with a nontargeting negative control pool, transfection reagent alone, or no treatment. (C) Comparison of individual siRNA pools targeting ZAP, ISG20, ISG15, or ISG56 with combinations of two siRNA pools and control treatments described for panel A above.
Similar articles
- Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells.
Ryman KD, Meier KC, Nangle EM, Ragsdale SL, Korneeva NL, Rhoads RE, MacDonald MR, Klimstra WB. Ryman KD, et al. J Virol. 2005 Feb;79(3):1487-99. doi: 10.1128/JVI.79.3.1487-1499.2005. J Virol. 2005. PMID: 15650175 Free PMC article. - Alpha/beta interferon inhibits cap-dependent translation of viral but not cellular mRNA by a PKR-independent mechanism.
Tesfay MZ, Yin J, Gardner CL, Khoretonenko MV, Korneeva NL, Rhoads RE, Ryman KD, Klimstra WB. Tesfay MZ, et al. J Virol. 2008 Mar;82(6):2620-30. doi: 10.1128/JVI.01784-07. Epub 2007 Dec 26. J Virol. 2008. PMID: 18160435 Free PMC article. - Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis.
Ryman KD, White LJ, Johnston RE, Klimstra WB. Ryman KD, et al. Viral Immunol. 2002;15(1):53-76. doi: 10.1089/088282402317340233. Viral Immunol. 2002. PMID: 11952147 - Interferon-Stimulated Genes that Target Retrovirus Translation.
Jäger N, Pöhlmann S, Rodnina MV, Ayyub SA. Jäger N, et al. Viruses. 2024 Jun 8;16(6):933. doi: 10.3390/v16060933. Viruses. 2024. PMID: 38932225 Free PMC article. Review. - ISG20: an enigmatic antiviral RNase targeting multiple viruses.
Deymier S, Louvat C, Fiorini F, Cimarelli A. Deymier S, et al. FEBS Open Bio. 2022 Jun;12(6):1096-1111. doi: 10.1002/2211-5463.13382. Epub 2022 Feb 27. FEBS Open Bio. 2022. PMID: 35174977 Free PMC article. Review.
Cited by
- Neurotropic arboviruses induce interferon regulatory factor 3-mediated neuronal responses that are cytoprotective, interferon independent, and inhibited by Western equine encephalitis virus capsid.
Peltier DC, Lazear HM, Farmer JR, Diamond MS, Miller DJ. Peltier DC, et al. J Virol. 2013 Feb;87(3):1821-33. doi: 10.1128/JVI.02858-12. Epub 2012 Nov 28. J Virol. 2013. PMID: 23192868 Free PMC article. - Multiple interferon stimulated genes synergize with the zinc finger antiviral protein to mediate anti-alphavirus activity.
Karki S, Li MM, Schoggins JW, Tian S, Rice CM, MacDonald MR. Karki S, et al. PLoS One. 2012;7(5):e37398. doi: 10.1371/journal.pone.0037398. Epub 2012 May 16. PLoS One. 2012. PMID: 22615998 Free PMC article. - The broad-spectrum antiviral functions of IFIT and IFITM proteins.
Diamond MS, Farzan M. Diamond MS, et al. Nat Rev Immunol. 2013 Jan;13(1):46-57. doi: 10.1038/nri3344. Epub 2012 Dec 14. Nat Rev Immunol. 2013. PMID: 23237964 Free PMC article. Review. - Distinct Cellular Tropism and Immune Responses to Alphavirus Infection.
Kafai NM, Diamond MS, Fox JM. Kafai NM, et al. Annu Rev Immunol. 2022 Apr 26;40:615-649. doi: 10.1146/annurev-immunol-101220-014952. Epub 2022 Feb 8. Annu Rev Immunol. 2022. PMID: 35134315 Free PMC article. Review. - Cell-type- and region-specific restriction of neurotropic flavivirus infection by viperin.
Lindqvist R, Kurhade C, Gilthorpe JD, Överby AK. Lindqvist R, et al. J Neuroinflammation. 2018 Mar 15;15(1):80. doi: 10.1186/s12974-018-1119-3. J Neuroinflammation. 2018. PMID: 29544502 Free PMC article.
References
- Anderson, S. L., J. M. Carton, J. Lou, L. Xing, and B. Y. Rubin. 1999. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256:8-14. - PubMed
- Anishchenko, M., S. Paessler, I. P. Greene, P. V. Aguilar, A. S. Carrara, and S. C. Weaver. 2004. Generation and characterization of closely related epizootic and enzootic infectious cDNA clones for studying interferon sensitivity and emergence mechanisms of Venezuelan equine encephalitis virus. J. Virol. 78:1-8. - PMC - PubMed
- Boxel-Dezaire, A. H., M. R. Rani, and G. R. Stark. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25:361-372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AI22186/AI/NIAID NIH HHS/United States
- AI069158-01/AI/NIAID NIH HHS/United States
- R21 AI069158/AI/NIAID NIH HHS/United States
- P20 RR 018724-01/RR/NCRR NIH HHS/United States
- R01 AI022186/AI/NIAID NIH HHS/United States
- P20 RR018724/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous