Application of a BRAF pyrosequencing assay for mutation detection and copy number analysis in malignant melanoma - PubMed (original) (raw)
Application of a BRAF pyrosequencing assay for mutation detection and copy number analysis in malignant melanoma
Cynthia Spittle et al. J Mol Diagn. 2007 Sep.
Abstract
Mutations in the BRAF gene are found in the majority of cutaneous malignant melanomas and subsets of other tumors. These mutations lead to constitutive activation of BRAF with increased downstream ERK (extracellular signal-regulated kinase) signaling; therefore, the development of RAF kinase inhibitors for targeted therapy is being actively pursued. A methodology that allows sensitive, cost-effective, high-throughput analysis of BRAF mutations will be needed to triage patients for specific molecular-based therapies. Pyrosequencing is a high-throughput, sequencing-by-synthesis method that is particularly useful for analysis of single nucleotide polymorphisms or hotspot mutations. Mutational analysis of BRAF is highly amenable to pyrosequencing because the majority of mutations in this gene localize to codons 600 and 601 and consist of single or dinucleotide substitutions. In this study, DNAs from a panel of melanocyte cell lines, melanoma cell lines, and melanoma tumors were used to validate a pyrosequencing assay to detect BRAF mutations. The assay demonstrates high accuracy and precision for detecting common and variant exon 15 BRAF mutations. Further, comparison of pyrosequencing data with 100K single nucleotide polymorphism microarray data allows characterization of BRAF amplification events that may accompany BRAF mutation. Pyro-sequencing serves as an excellent platform for BRAF genotyping of tumors from patients entering clinical trial.
Figures
Figure 1
Detection of V600E in melanoma cell lines. Pyrograms generated for BRAF wild-type (WM3208) (A), heterozygous mutant (WM 902B) (B), and homozygous mutant (WM39) (C) cell lines distinguish T versus A peaks (yellow shading). Nucleotide dispensation order is given below each pyrogram. Numerical position for each nucleotide is indicated at the top. Dispensations of C at positions 1 and 6 are included as controls for misincorporation. Dideoxy cycle sequence tracings for each cell line are shown for comparison.
Figure 2
Detection of BRAF variant mutations. Pyrograms for two variant mutations, V600R (B) and V600K (C), show distinct patterns compared with V600E (A). Changes in relative A/G peak heights at pyrogram positions 2, 3, 5, 7, and/or 8 reflect dinucleotide substitutions given at right. The specific variants are confirmed by dideoxy cycle sequencing.
Figure 3
Applicability of pyrosequencing assay to FFPE tissue. Pyrograms generated from a V600E mutant tumor (A) and a V600K mutant tumor (B) are identical for paired frozen and FFPE tumors.
Figure 4
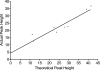
Cell line DNA mixing study. Theoretical peak heights, calculated from initial percent mutant A peak in undiluted heterozygous V600E mutant tumor, are correlated with actual peak heights generated for each dilution (Pearson’s correlation = 0.96), indicating a linear relationship.
Figure 5
Analytic sensitivity for V600E detection in a heterozygous tumor. Pyrograms generated from dilution series for two separate experiments (day 1 and day 2) are shown with corresponding dideoxy sequence tracings for day 1 data. The percent tumor corresponding to each data set is indicated at left.
Figure 6
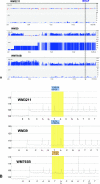
Comparison of pyrosequencing data with SNP microarrays. A: SNP _Xba_I 50K microarray data for chromosome 7 displayed with CNAT 3.0 software. Melanoma cell lines analyzed and location of BRAF gene are indicated. For each cell line, genome smoothed chromosome copy number data are shown at the top and the LOH score is shown at the bottom. Chromosome location in megabases is given at the bottom of the data set for each cell line. Scale for copy number and LOH score indicated at right. B: Corresponding BRAF pyrosequencing data including percent T and A peaks are indicated for each cell line.
Similar articles
- Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis.
Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Willmore-Payne C, et al. Hum Pathol. 2005 May;36(5):486-93. doi: 10.1016/j.humpath.2005.03.015. Hum Pathol. 2005. PMID: 15948115 - BRAF and c-kit gene copy number in mutation-positive malignant melanoma.
Willmore-Payne C, Holden JA, Hirschowitz S, Layfield LJ. Willmore-Payne C, et al. Hum Pathol. 2006 May;37(5):520-7. doi: 10.1016/j.humpath.2006.01.003. Hum Pathol. 2006. PMID: 16647948 - Next-Generation Genotyping by Digital PCR to Detect and Quantify the BRAF V600E Mutation in Melanoma Biopsies.
Lamy PJ, Castan F, Lozano N, Montélion C, Audran P, Bibeau F, Roques S, Montels F, Laberenne AC. Lamy PJ, et al. J Mol Diagn. 2015 Jul;17(4):366-73. doi: 10.1016/j.jmoldx.2015.02.004. Epub 2015 May 5. J Mol Diagn. 2015. PMID: 25952101 - BRAF Mutation Status Concordance Between Primary Cutaneous Melanomas and Corresponding Metastases: A Review of the Latest Evidence.
Godoy-Gijón E, Yuste-Chaves M, Santos-Briz Á. Godoy-Gijón E, et al. Actas Dermosifiliogr. 2017 Dec;108(10):894-901. doi: 10.1016/j.ad.2016.12.025. Epub 2017 Jul 12. Actas Dermosifiliogr. 2017. PMID: 28711165 Review. English, Spanish. - New insight into BRAF mutations in cancer.
Dhomen N, Marais R. Dhomen N, et al. Curr Opin Genet Dev. 2007 Feb;17(1):31-9. doi: 10.1016/j.gde.2006.12.005. Curr Opin Genet Dev. 2007. PMID: 17208430 Review.
Cited by
- CXCR4 pathway associated with family history of melanoma.
Li WQ, Han J, Widlund HR, Correll M, Wang YE, Quackenbush J, Mihm MC, Canales AL, Wu S, Golub T, Hoshida Y, Hunter DJ, Murphy G, Kupper TS, Qureshi AA. Li WQ, et al. Cancer Causes Control. 2014 Jan;25(1):125-32. doi: 10.1007/s10552-013-0315-9. Epub 2013 Oct 25. Cancer Causes Control. 2014. PMID: 24158781 Free PMC article. - BRAFV600E immunopositive melanomas show low frequency of heterogeneity and association with epithelioid tumor cells: a STROBE-compliant article.
Verlinden I, van den Hurk K, Clarijs R, Willig AP, Stallinga CMHA, Roemen GMJM, van den Oord JJ, Zur Hausen A, Speel EM, Winnepenninckx VJL. Verlinden I, et al. Medicine (Baltimore). 2014 Dec;93(28):e285. doi: 10.1097/MD.0000000000000285. Medicine (Baltimore). 2014. PMID: 25526463 Free PMC article. - BRAF pyrosequencing analysis aided by a lookup table.
Olson MT, Harrington C, Beierl K, Chen G, Thiess M, O'Neill A, Taube JM, Zeiger MA, Lin MT, Eshleman JR. Olson MT, et al. Am J Clin Pathol. 2014 May;141(5):639-47. doi: 10.1309/AJCPVWH1K2ZIHHTV. Am J Clin Pathol. 2014. PMID: 24713734 Free PMC article. - BRAF-mutant melanoma: treatment approaches, resistance mechanisms, and diagnostic strategies.
Spagnolo F, Ghiorzo P, Orgiano L, Pastorino L, Picasso V, Tornari E, Ottaviano V, Queirolo P. Spagnolo F, et al. Onco Targets Ther. 2015 Jan 16;8:157-68. doi: 10.2147/OTT.S39096. eCollection 2015. Onco Targets Ther. 2015. PMID: 25653539 Free PMC article. Review. - Rare BRAF mutations in melanoma patients: implications for molecular testing in clinical practice.
Heinzerling L, Kühnapfel S, Meckbach D, Baiter M, Kaempgen E, Keikavoussi P, Schuler G, Agaimy A, Bauer J, Hartmann A, Kiesewetter F, Schneider-Stock R. Heinzerling L, et al. Br J Cancer. 2013 May 28;108(10):2164-71. doi: 10.1038/bjc.2013.143. Epub 2013 Apr 11. Br J Cancer. 2013. PMID: 23579220 Free PMC article.
References
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. - PubMed
- Cohen Y, Xing M, Mambo E, Guo Z, Wu G, Trink B, Beller U, Westra WH, Ladenson PW, Sidransky D. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. - PubMed
- Singer G, Oldt R, III, Cohen Y, Wang BG, Sidransky D, Kurman RJ, Shih Ie M. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. - PubMed
- Yuen ST, Davies H, Chan TL, Ho JW, Bignell GR, Cox C, Stephens P, Edkins S, Tsui WW, Chan AS, Futreal PA, Stratton MR, Wooster R, Leung SY. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62:6451–6455. - PubMed
- Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous