Cell- and gene-specific regulation of primary target genes by the androgen receptor - PubMed (original) (raw)
Cell- and gene-specific regulation of primary target genes by the androgen receptor
Eric C Bolton et al. Genes Dev. 2007.
Abstract
The androgen receptor (AR) mediates the physiologic and pathophysiologic effects of androgens including sexual differentiation, prostate development, and cancer progression by binding to genomic androgen response elements (AREs), which influence transcription of AR target genes. The composition and context of AREs differ between genes, thus enabling AR to confer multiple regulatory functions within a single nucleus. We used expression profiling of an immortalized human prostate epithelial cell line to identify 205 androgen-responsive genes (ARGs), most of them novel. In addition, we performed chromatin immunoprecipitation to identify 524 AR binding regions and validated in reporter assays the ARE activities of several such regions. Interestingly, 67% of our AREs resided within approximately 50 kb of the transcription start sites of 84% of our ARGs. Indeed, most ARGs were associated with two or more AREs, and ARGs were sometimes themselves linked in gene clusters containing up to 13 AREs and 12 ARGs. AREs appeared typically to be composite elements, containing AR binding sequences adjacent to binding motifs for other transcriptional regulators. Functionally, ARGs were commonly involved in prostate cell proliferation, communication, differentiation, and possibly cancer progression. Our results provide new insights into cell- and gene-specific mechanisms of transcriptional regulation of androgen-responsive gene networks.
Figures
Figure 1.
qPCR validation of the expression of ARGs identified by microarray. (A) Most of the 157 putative ARGs identified by expression microarray analysis were subsequently validated using qPCR. The colorimetric representation shows genes, indicated by HUGO Gene Nomenclature Committee gene symbols, whose transcripts were repressed (B, green) and induced (C, red) by androgen. The color intensity reflects the relative fold change in transcript level for androgen- versus vehicle-treated cells. The time course spanned 4, 8, 15, and 24 h. The mean change in expression level for genes above the orange line was greater than twofold across the time course, whereas the mean change in expression for those genes below the line was greater than 1.5 fold.
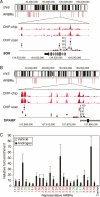
Figure 2.
ARBRs: identification by ChIP–chip and validation by conventional ChIP. The visual representation of ARBRs on chromosomes 6 (A) and 3 (B) are shown. ARBRs are shown as equilateral red bars immediately below the chromosomes. Expanded views of (A) SGK and (B) TIPARP gene loci are shown based on the May 2004 human genome freeze hg17 in the UCSC browser using GenBank RefSeq positions. For SGK (3′–5′ orientation) and TIPARP (5′–3′ orientation), the ChIP–chip signals (red bars) are shown for duplicate experiments and indicate the relative fold enrichment of AR-immunoprecipitated DNA fragments for androgen- versus vehicle-treated cells. The ChIP scanning (Wang et al. 2004) signals (red circles) demonstrate validation of the ChIP–chip data and show the relative fold enrichment of AR ChIP for androgen- versus vehicle-treated cells. Data represent the mean ± SD, n = 4. (C) Additional ARBRs (numbered) identified by ChIP–chip were validated using conventional AR ChIP. The relative fold enrichment of AR ChIP is shown for androgen- versus vehicle-treated cells; data represent the mean ± SD, n = 4. The ARBRs found near or within genes that are repressed (green), induced (red), or unresponsive (black) following androgen treatment are indicated. A region near the HSPA1A gene, which is not occupied by AR, is shown as a control. Compared to the control, all ARBRs demonstrate significant AR occupancy, P < 0.05.
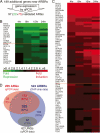
Figure 3.
ARBRs identified by AR ChIP-chip lead to the discovery of novel primary ARGs. (A) Many of the 189 ARBR-proximal genes that were not detected by microarray analysis were, in fact, primary ARGs. The colorimetric representation shows genes, near identified ARBRs, whose transcripts were repressed (B, green) or induced (C, red) by androgen treatment. See Figure 1 for colorimetric and time course details. (D) Of the 524 ARBRs (blue) and 205 (108 initial and 97 additional ARGs, red) differentially expressed ARGs, we found 352 ARBRs near 172 ARGs, which represent direct, functional targets of AR. The response of 62 genes containing in total 68 intragenic ARBRs (gray) was independent of androgen (androgen-unresponsive genes, URGs).
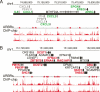
Figure 4.
Intersection of ARBR and ARG data reveals primary ARG clusters. (A) An 800-kb region on chromosome 4 containing seven validated ARBRs (red asterisks) was found to contain genes that were repressed (green) or unresponsive (black) following treatment with androgen. (B) A 370-kb region on chromosome 1 containing 13 validated ARBRs (red asterisks) was found to contain genes that were induced (red) or unresponsive (black) following treatment with androgen. Gene loci are shown based on the May 2004 human genome freeze hg17 in the UCSC browser using GenBank RefSeq positions. The androgen responsiveness of some genes was not determined (gray). Genes above the heavy black line are transcribed from left to right whereas those below the line are transcribed from right to left. The longest transcriptional variants for each gene are represented. The ChIP–chip signals (red bars) are shown for duplicate experiments and indicate the relative fold enrichment of AR-immunoprecipitated DNA fragments for androgen- versus vehicle-treated cells.
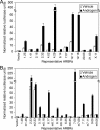
Figure 5.
ARBRs display androgen response element (ARE) activities. (A) Thirteen sequences of ∼500 bp, each containing individual ARBRs (numbered), were cloned upstream of a minimal promoter sequence driving the expression of luciferase. (B) Nine additional sequences of ∼500 bp, each containing individual ARBRs (numbered), were also cloned as described (see Materials and Methods). These nine ARBRs and the corresponding ARBR mutants, which contain mutated ARBSs (indicated by the letter m preceding the ARBR number), were tested in cell-based reporter assays. HPr-1AR cells transfected with each of the ARBR reporter constructs were treated with vehicle (open bars) or androgen R1881 (solid bars) and subsequently were assayed for androgen-stimulated luciferase activity. The ARBRs shown correspond to sequences upstream of, within, and downstream from ARGs. Data represent the mean ± SD, n = 3.
Figure 6.
Distribution analysis of AREs located near ARGs. (A) The distribution of AREs identified by ChIP–chip was plotted relative to the predicted transcription start site (+1) of the longest transcriptional variant of the nearest ARG. AREs reside great distances upstream and downstream, as well as near the promoter. Notably, fewer AREs were found upstream of than downstream from ARG transcription start sites of the longest transcriptional variants. Relative distances upstream of (negative) and downstream (positive) from the transcription start sites (vertical black line) were assigned to 10 kb bins. AREs were generally found beyond 10 kb of the transcription start site. (B) The location of AREs is shown relative to the predicted gene substructure of the longest transcriptional variant of the nearby ARGs. When the distribution of all AREs near all ARGs was assessed, most AREs were situated downstream from the transcription start site. These distributions were subdivided based on whether the ARG was repressed or induced, suggesting biases in terms of ARE location and the mean AREs/ARG ratios.
Figure 7.
ARBSs and other _cis_-regulatory motifs are enriched within AREs. Unbiased motif searches of all 524 AREs revealed the presence of significantly recurring motifs. (A) BioProspector analysis identified a consensus ARBS, which is visually represented in WebLogo (
). (B) MobyDick analysis detected a consensus AR half-site motif as well as putative motifs for AP-1, RAR, ZNF42, HNF-4α, and EGR. These enriched sequence motifs are represented using IUPAC symbols (
). Motifs where the putative regulatory factors were not determined (ND) are indicated. _P_-value calculations are described in Materials and Methods.
Similar articles
- Identification and functional analysis of consensus androgen response elements in human prostate cancer cells.
Horie-Inoue K, Bono H, Okazaki Y, Inoue S. Horie-Inoue K, et al. Biochem Biophys Res Commun. 2004 Dec 24;325(4):1312-7. doi: 10.1016/j.bbrc.2004.10.174. Biochem Biophys Res Commun. 2004. PMID: 15555570 - Two androgen response elements in the androgen receptor coding region are required for cell-specific up-regulation of receptor messenger RNA.
Dai JL, Burnstein KL. Dai JL, et al. Mol Endocrinol. 1996 Dec;10(12):1582-94. doi: 10.1210/mend.10.12.8961268. Mol Endocrinol. 1996. PMID: 8961268 - Molecular regulation of androgen action in prostate cancer.
Dehm SM, Tindall DJ. Dehm SM, et al. J Cell Biochem. 2006 Oct 1;99(2):333-44. doi: 10.1002/jcb.20794. J Cell Biochem. 2006. PMID: 16518832 Review. - Androgen receptor enhancer usage and the chromatin regulatory landscape in human prostate cancers.
Stelloo S, Bergman AM, Zwart W. Stelloo S, et al. Endocr Relat Cancer. 2019 May;26(5):R267-R285. doi: 10.1530/ERC-19-0032. Endocr Relat Cancer. 2019. PMID: 30865928 Review.
Cited by
- Androgen Receptor-dependent Expression of Low-density Lipoprotein Receptor-related Protein 6 is Necessary for Prostate Cancer Cell Proliferation.
Park E, Kim EK, Kim M, Ha JM, Kim YW, Jin SY, Shin HK, Ha HK, Lee JZ, Bae SS. Park E, et al. Korean J Physiol Pharmacol. 2015 May;19(3):235-40. doi: 10.4196/kjpp.2015.19.3.235. Epub 2015 Apr 30. Korean J Physiol Pharmacol. 2015. PMID: 25954128 Free PMC article. - Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions.
Decker KF, Zheng D, He Y, Bowman T, Edwards JR, Jia L. Decker KF, et al. Nucleic Acids Res. 2012 Nov;40(21):10765-79. doi: 10.1093/nar/gks888. Epub 2012 Sep 27. Nucleic Acids Res. 2012. PMID: 23019221 Free PMC article. - Regulation of several androgen-induced genes through the repression of the miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells.
Sun D, Layer R, Mueller AC, Cichewicz MA, Negishi M, Paschal BM, Dutta A. Sun D, et al. Oncogene. 2014 Mar 13;33(11):1448-57. doi: 10.1038/onc.2013.77. Epub 2013 Mar 18. Oncogene. 2014. PMID: 23503464 Free PMC article. - Salvia miltiorrhiza Bunge Ameliorates Benign Prostatic Hyperplasia through Regulation of Oxidative Stress via Nrf-2/HO-1 Activation.
Choi YJ, Wedamulla NE, Kim SH, Oh M, Seo KS, Han JS, Lee EJ, Park YH, Park YJ, Kim EK. Choi YJ, et al. J Microbiol Biotechnol. 2024 May 28;34(5):1059-1072. doi: 10.4014/jmb.2308.08053. Epub 2023 Oct 31. J Microbiol Biotechnol. 2024. PMID: 37994101 Free PMC article. - Glucocorticoid regulation of the circadian clock modulates glucose homeostasis.
So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. So AY, et al. Proc Natl Acad Sci U S A. 2009 Oct 13;106(41):17582-7. doi: 10.1073/pnas.0909733106. Epub 2009 Oct 5. Proc Natl Acad Sci U S A. 2009. PMID: 19805059 Free PMC article.
References
- Belperio J.A., Keane M.P., Arenberg D.A., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Keane M.P., Arenberg D.A., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Arenberg D.A., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Ehlert J.E., Burdick M.D., Strieter R.M., Burdick M.D., Strieter R.M., Strieter R.M. CXC chemokines in angiogenesis. J. Leukoc. Biol. 2000;68:1–8. - PubMed
- Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Shao W., Hestermann E.V., Geistlinger T.R., Hestermann E.V., Geistlinger T.R., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. - PubMed
- Chang C.S., Kokontis J., Liao S.T., Kokontis J., Liao S.T., Liao S.T. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science. 1988;240:324–326. - PubMed
- Chang T.Y., Chang C.C., Lin S., Yu C., Li B.L., Miyazaki A., Chang C.C., Lin S., Yu C., Li B.L., Miyazaki A., Lin S., Yu C., Li B.L., Miyazaki A., Yu C., Li B.L., Miyazaki A., Li B.L., Miyazaki A., Miyazaki A. Roles of acyl-coenzyme A:cholesterol acyltransferase-1 and -2. Curr. Opin. Lipidol. 2001;12:289–296. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM070808/GM/NIGMS NIH HHS/United States
- R01 CA020535/CA/NCI NIH HHS/United States
- CA020535/CA/NCI NIH HHS/United States
- F32DK65402/DK/NIDDK NIH HHS/United States
- R01 CA101042/CA/NCI NIH HHS/United States
- GM070808/GM/NIGMS NIH HHS/United States
- CA101042/CA/NCI NIH HHS/United States
- R37 CA020535/CA/NCI NIH HHS/United States
- F32 DK065402/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials