Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation - PubMed (original) (raw)
Comparative Study
Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation
Sanjeev Das et al. Cell. 2007.
Abstract
A critical unresolved issue about the genotoxic stress response is how the resulting activation of the p53 tumor suppressor can lead either to cell-cycle arrest and DNA repair or to apoptosis. We show here that hematopoietic zinc finger (Hzf), a zinc-finger-containing p53 target gene, modulates p53 transactivation functions in an autoregulatory feedback loop. Hzf is induced by p53 and binds to its DNA-binding domain, resulting in preferential transactivation of proarrest p53 target genes over its proapoptotic target genes. Thus, p53 activation results in cell-cycle arrest in Hzf wild-type MEFs, while in Hzf(-/-) MEFs, apoptosis is induced. Exposure of Hzf null mice to ionizing radiation resulted in enhanced apoptosis in several organs, as compared to in wild-type mice. These findings provide novel insights into the regulation of p53 transactivation function and suggest that Hzf functions as a key player in regulating cell fate decisions in response to genotoxic stress.
Figures
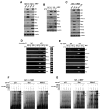
Figure 1. Hzf induction in response to various stresses modulates p53 transcriptional function
(A) Hzf is induced by genotoxic and oxidative stress in a p53-dependent manner. HCT116, HCT116 p53−/−, U2OS and LNCaP cells were treated with etoposide (ETO, 40 μM), IR (4 Gy) or H2O2 (0.4 mM) for the indicated time points. The cells were then harvested, and northern blots or western blots were carried out. (B) Loss of Hzf represses p21 induction but enhances Bax expression in response to DNA damage or ectopic p53. U2OS and EJ cells were treated with etoposide (ETO, 40 μM) or infected with Ad-p53 respectively for 24 hours. The cells were then harvested and Western blots carried out for the indicated proteins. (C) Wt and Hzf−/− MEFs were treated with etoposide (ETO, 40 μM) for 24 hours. The cells were then harvested and Northern blots carried out as indicated. (D) Effects of Hzf inhibition on p53-mediated transcription. Reporter gene constructs containing the p53-responsive elements (pGL3-p21-Luc or pGL3-Bax-Luc) or control vector alone (pGL3-Basic) were co-transfected into U2OS and Saos2 cells with either Hzf shRNA or control scrambled shRNA, and 24 hours later cells were treated with etoposide or infected with Ad-p53 for another 24 hours. At the end of the time point Luciferase assay was carried out. Error bars are means ± SD of three independent experiments with duplicate samples.
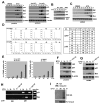
Figure 2. Hzf physically interacts with p53
(A) U2OS and EJ cells were treated with ETO (40 μM) or infected with Ad-p53 respectively for 24 hours. The cells were then harvested and subjected to immunoprecipitations using anti-p53 antibody (top panels) or anti-Hzf antibody (bottom panels) and Western blots carried out for the indicated proteins. (B) Yeast two hybrid assay to show direct interaction between Hzf and p53 was carried out as described in methods. (C) The region of p53 involved in binding to Hzf was analyzed in GST-pull down assays. U2OS cells were transfected with the indicated GST-p53 plasmids and then infected with Ad-GFP or Ad-Hzf. 24 hours post-infection the cells were lysed and subjected to GST pull down. Western blotting was then carried out for p53, and Flag epitope. (D) Mutations in DNA binding domain of p53 affect the Hzf-p53 interaction. EJ cells containing unfunctional p53 were transfected with the indicated p53 plasmids and then infected with Ad-GFP or Ad-Hzf. 24 hours post-infection the cells were lysed and subjected to immunoprecipitations using anti-p53 antibody. Western blotting was carried out for p53, and Flag epitope.
Figure 3. p53 bound to Hzf is preferentially recruited to p21 and 14-3-3σ promoter
(A, B) Loss of Hzf enhances expression of proapoptotic targets of p53, but it suppresses the expression of p21 and 14-3-3σ. Western blotting of p53 targets in Hzf+/+ and Hzf−/−MEFs. Hzf+/+ and Hzf−/− MEFs were infected with Ad-GFP or Ad-p53 (A) or treated with ETO (40 μM) (B). 24 hours later, Western blot analysis was carried out for the indicated proteins. (C) Re-expression of Hzf restores preferential induction of p21 and 14-3-3σ and represses proapoptotic targets in response to p53. Hzf−/− MEFs were infected with Ad-GFP or Ad-Hzf 6 hours prior to infection with Ad-GFP or Ad-p53. 24 hours post infection, Western blots were carried out for the indicated proteins. (D) Multiple ChIP analysis on the promoters of p53 targets. Wt and Hzf−/− MEFs were treated with ETO (40 μM). 24 hours post drug treatment ChIP was carried out using control mouse IgG or anti-p53 antibody and PCR was done for the indicate promoters. (E) Part of the chromatin immunoprecipitated with p53 antibody was again subjected to ChIP using control rabbit IgG or Hzf antibody. Input represents 2% of total chromatin from untreated Hzf+/+ MEFs used for immunoprecipitation. (F, G) EMSA showing the DNA binding activity of p53 to oligonucleotides containing the p53 binding site in p21 promoter, Bax first intron, and Mdm2 promoter in wt-MEF (F) and Hzf-null MEF (G). Wt and Hzf−/− MEFs were treated with ETO (40 μM). 24 hours post drug treatment, nuclear extracts were prepared, and EMSA carried out as described in supplementary methods. The arrows indicate the position of the p53-DNA complex (lower) and supershifted complex containing anti-p53 antibody (pAb421).
Figure 4. Hzf promotes cell cycle arrest function of p53
(A) Hzf+/+ and Hzf−/− MEFs were infected with Ad-GFP or Ad-p53. 24 hours post-infection FACS analysis was carried out. (B) Hzf+/+ and Hzf−/− MEFs were similarly treated as in (A) and TUNEL staining and DNA fragmentation assays were carried out. Error bars are means ± SD of three independent experiments. (C) Re-expression of Hzf decreases apoptosis in response to ETO or p53 overexpression in Hzf−/− MEFs. Wt-MEFs and Hzf−/− MEFs were treated with ETO (40 μM). In cases where the cells were infected with Ad-GFP or Ad-Hzf prior to drug treatment, there was a difference of 6 hours between viral infection and drug treatment. 24 hours post drug treatment the cells were subjected to TUNEL assay. Error bars are means ± SD of three independent experiments.
Figure 5. Sustained p53 activation promotes ubiquitination and degradation of Hzf
(A) Prolonged p53 activation induces Hzf protein down-regulation. U2OS cells were exposed to ETO (40 μM) or were infected with Ad-GFP or Ad-p53 for the indicated time points. The cells were lysed and Western blots were carried out for the indicated proteins. (B) U2OS cells exposed to ETO (40 μM) for the indicated time points were immunoprecipitated with p53 Ab, and the precipitates were immunoblotted with Hzf Ab. (C) Hzf mRNA levels in response to ETO treatment remain steady. U2OS cells were exposed to ETO (40 μM) for the indicated time points and Northern blots were carried out as indicated. (D) U2OS cells were infected with Ad-GFP or Ad-p53 for the indicated time points and FACS analysis was carried out. (E) U2OS cells were similarly treated as in (A) and TUNEL assays were carried out for the indicated time points. Error bars are means ± SD of three independent experiments. (F, G, H) Proteasome-dependent Hzf degradation. U2OS cells were treated for 36 and 72 hours with ETO (40 μM), and MG132 (10 μM) was added during the last 10 hours prior to the end of the 72 hours time point. The cells were then harvested, and Western blotting, Northern blotting or ChIP was carried out with the indicated antibodies. Input represents 2% of total chromatin from untreated U2OS cells used for immunoprecipitation. (I) Genotoxic stress-induced ubiquitination of Hzf. U2OS cells were treated with ETO (40 μM) for 36 and 72 hours, and with MG132 for the last 10 hours. Cell extracts were immunoprecipitated with anti-Hzf Ab, and immunoblotted with anti-ubiquitin Ab.
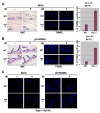
Figure 6. Increased apoptosis after radiation-induced DNA damage in skin and prostate of Hzf-deficient mice versus wild-type mice
(A) Representative TUNEL staining of skin from Hzf−/− mice and wild-type littermates, analyzed 6.5 hours after the mice received 5 Gy total-body irradiation. Representative H&E sections are shown (left panel). Percentages of TUNEL positive cells were calculated from three independent experiments (right panel). (B) Representative TUNEL staining of prostate tissue from the same IR treated Hzf−/−mice and wild-type littermates as in (A). Representative H&E sections are shown (left panel). Percentage of TUNEL positive cells were calculated from three independent experiments (right panel). (C) Immunostaining for Hzf in skin and prostate from the same IR treated Hzf−/− mice and wild-type littermates as in Figure 6A.
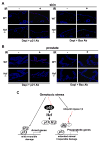
Figure 7. Loss of Hzf enhances the expression of Bax, but the levels of p21 expression are reduced in skin and prostate
(A) Immunostaining for p21 and Bax proteins in skin from the same IR treated Hzf−/− mice and wild-type littermates as in Figure 6A. (B) Immunostaining for p21 and Bax proteins in prostate tissue from the same IR treated Hzf−/− mice and wild-type littermates as in Figure 6B. (C) Model for p53-dependent cell fate control through the p53→Hzf→p53/Hzf autoregulatory feedback loop. p53 activation by DNA damage induces Hzf, and it binds to p53. This directs p53 preferentially to cell cycle arrest gene promoters resulting in growth arrest. Upon extended or irreparable stress, Hzf ubiquitination/degradation prevents this from occurring, thus allowing p53 to activate pro-apoptotic targets, and resulting in cell to trigger apoptosis.
Comment in
- Living with p53, dying of p53.
Aylon Y, Oren M. Aylon Y, et al. Cell. 2007 Aug 24;130(4):597-600. doi: 10.1016/j.cell.2007.08.005. Cell. 2007. PMID: 17719538 Review.
Similar articles
- PGC-1α, a key modulator of p53, promotes cell survival upon metabolic stress.
Sen N, Satija YK, Das S. Sen N, et al. Mol Cell. 2011 Nov 18;44(4):621-34. doi: 10.1016/j.molcel.2011.08.044. Mol Cell. 2011. PMID: 22099309 - Che-1 modulates the decision between cell cycle arrest and apoptosis by its binding to p53.
Desantis A, Bruno T, Catena V, De Nicola F, Goeman F, Iezzi S, Sorino C, Gentileschi MP, Germoni S, Monteleone V, Pellegrino M, Kann M, De Meo PD, Pallocca M, Höpker K, Moretti F, Mattei E, Reinhardt HC, Floridi A, Passananti C, Benzing T, Blandino G, Fanciulli M. Desantis A, et al. Cell Death Dis. 2015 May 21;6(5):e1764. doi: 10.1038/cddis.2015.117. Cell Death Dis. 2015. PMID: 25996291 Free PMC article. - HDAC5, a key component in temporal regulation of p53-mediated transactivation in response to genotoxic stress.
Sen N, Kumari R, Singh MI, Das S. Sen N, et al. Mol Cell. 2013 Nov 7;52(3):406-20. doi: 10.1016/j.molcel.2013.09.003. Epub 2013 Oct 10. Mol Cell. 2013. PMID: 24120667 - The role of p53 as a transcription factor in the induction of apoptosis.
Yonish-Rouach E, Choisy C, Deguin V, Breugnot C, May E. Yonish-Rouach E, et al. Behring Inst Mitt. 1996 Oct;(97):60-71. Behring Inst Mitt. 1996. PMID: 8950467 Review. - [Relationships between p53 induction, cell cycle arrest and survival of normal human fibroblasts following DNA damage].
Ceraline J, Deplanque G, Duclos B, Limacher JM, Vincent F, Goldblum S, Bergerat JP. Ceraline J, et al. Bull Cancer. 1997 Nov;84(11):1007-16. Bull Cancer. 1997. PMID: 9536982 Review. French.
Cited by
- Cell-to-Cell Variation in p53 Dynamics Leads to Fractional Killing.
Paek AL, Liu JC, Loewer A, Forrester WC, Lahav G. Paek AL, et al. Cell. 2016 Apr 21;165(3):631-42. doi: 10.1016/j.cell.2016.03.025. Epub 2016 Apr 7. Cell. 2016. PMID: 27062928 Free PMC article. - A distinct response to endogenous DNA damage in the development of Nbs1-deficient cortical neurons.
Li R, Yang YG, Gao Y, Wang ZQ, Tong WM. Li R, et al. Cell Res. 2012 May;22(5):859-72. doi: 10.1038/cr.2012.3. Epub 2012 Jan 3. Cell Res. 2012. PMID: 22212482 Free PMC article. - Transcriptomic analysis of paired healthy human skeletal muscles to identify modulators of disease severity in DMD.
Nieves-Rodriguez S, Barthélémy F, Woods JD, Douine ED, Wang RT, Scripture-Adams DD, Chesmore KN, Galasso F, Miceli MC, Nelson SF. Nieves-Rodriguez S, et al. Front Genet. 2023 Jul 27;14:1216066. doi: 10.3389/fgene.2023.1216066. eCollection 2023. Front Genet. 2023. PMID: 37576554 Free PMC article. - Formation of stress-specific p53 binding patterns is influenced by chromatin but not by modulation of p53 binding affinity to response elements.
Millau JF, Bandele OJ, Perron J, Bastien N, Bouchard EF, Gaudreau L, Bell DA, Drouin R. Millau JF, et al. Nucleic Acids Res. 2011 Apr;39(8):3053-63. doi: 10.1093/nar/gkq1209. Epub 2010 Dec 21. Nucleic Acids Res. 2011. PMID: 21177650 Free PMC article. - Interplay between Mdm2 and HIPK2 in the DNA damage response.
Zhang XP, Liu F, Wang W. Zhang XP, et al. J R Soc Interface. 2014 Jul 6;11(96):20140319. doi: 10.1098/rsif.2014.0319. J R Soc Interface. 2014. PMID: 24829283 Free PMC article.
References
- Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet. 2006;38:1133–1141. - PubMed
- Braithwaite AW, Del Sal G, Lu X. Some p53-binding proteins that can function as arbiters of life and death. Cell Death Differ. 2006;13:984–993. - PubMed
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1R01 CA097216/CA/NCI NIH HHS/United States
- R01 CA085214/CA/NCI NIH HHS/United States
- R01 CA078356-10/CA/NCI NIH HHS/United States
- P01 CA80058/CA/NCI NIH HHS/United States
- R01 CA085681-08/CA/NCI NIH HHS/United States
- R01 CA085681/CA/NCI NIH HHS/United States
- R01 CA097216/CA/NCI NIH HHS/United States
- 2R01 CA085681/CA/NCI NIH HHS/United States
- R01 CA097216-06/CA/NCI NIH HHS/United States
- P01 CA080058/CA/NCI NIH HHS/United States
- 2R01 CA078356/CA/NCI NIH HHS/United States
- 2R01 CA085214/CA/NCI NIH HHS/United States
- R01 CA078356/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous