New androgen receptor genomic targets show an interaction with the ETS1 transcription factor - PubMed (original) (raw)
New androgen receptor genomic targets show an interaction with the ETS1 transcription factor
Charles E Massie et al. EMBO Rep. 2007 Sep.
Abstract
The androgen receptor (AR) initiates important developmental and oncogenic transcriptional pathways. The AR is known to bind as a homodimer to 15-base pair bipartite palindromic androgen-response elements; however, few direct AR gene targets are known. To identify AR promoter targets, we used chromatin immunoprecipitation with on-chip detection of genomic fragments. We identified 1,532 potential AR-binding sites, including previously known AR gene targets. Many of the new AR target genes show altered expression in prostate cancer. Analysis of sequences underlying AR-binding sites showed that more than 50% of AR-binding sites did not contain the established 15 bp AR-binding element. Unbiased sequence analysis showed 6-bp motifs, which were significantly enriched and were bound directly by the AR in vitro. Binding sequences for the avian erythroblastosis virus E26 homologue (ETS) transcription factor family were also highly enriched, and we uncovered an interaction between the AR and ETS1 at a subset of AR promoter targets.
Figures
Figure 1
Androgen receptor-bound gene promoters identified by ChIP-chip. (A) AR ChIP-chip profiles of eight AR target gene promoters from vehicle (+EtOH)- and androgen (+R1881)-treated experiments. Genomic locations are indicated on the x axis and enrichment compared with total input DNA is shown on the y axis. (B) AR ChIP and qPCR analysis of 26 AR promoter targets in LNCaP cells. (C) AR ChIP and qPCR analysis of eight promoter targets in DUCaP cells. Relative enrichment by ChIP was quantified by using the standard curve method following 1 h with vehicle (EtOH) or androgen (R1881) relative to β-actin control and total genomic DNA input (see the supplementary information online for details). Values are the averages of replicate experiments. Enrichment scores (S) and Wilcoxon scores (W) from LNCaP AR ChIP-chip analysis are indicated next to corresponding genes. AR, androgen receptor; ChIP, chromatin immunoprecipitation; ChIP-chip, ChIP with on-array detection; Chr, chromosome; EtOH, ethyl alcohol; KLK, human glandular kellikrein; qPCR, quantitative PCR.
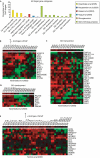
Figure 2
Functional annotation of androgen receptor ChIP target genes. (A) AR ChIP-chip target gene-enriched Gene Ontology categories and gene expression changes identified by comparing results with publicly available expression array data. (B–E) Heat-maps showing changes in gene expression for individual clinical samples, taken from publicly available expression array data (Varambally et al, 2005), for sets of genes identified as androgen-regulated (B), involved in transcription (C) and development (D). (E) Gene expression heat-map from a separate clinical expression array study for AR target genes that were identified as androgen-regulated (Tomlins et al, 2007). Arrows in B and E indicate that the expression of the genes selected have been reported to be upregulated on androgen treatment in previous studies. AR, androgen receptor; BN, benign; ChIP, chromatin immunoprecipitation; ChIP-chip, ChIP with on-array detection; EPI-BPH, benign prostatic hyperplasia; EPI-NORM, normal prostate epithelium; GO, Gene Ontology; KLK, human glandular kellikrein; MET, metastatic PrCa; PrCa, primary PrCa.
Figure 3
Sequence analysis of androgen receptor-bound promoter regions. (A) Sequence logo of the _in vitro_-derived 15-bp ARE (SELEX) and known genomic inverted repeat (IR) and direct repeat (DR) ARE sequences. (B) Venn diagram showing the occurrence of 15-bp AREs in AR ChIP-chip-enriched sequences. (C) Sequence logos of AR ‘half-site' motifs identified in AR-bound promoter sequences using Nested MICA. (D) Venn diagram of the co-occurrence of the 15-bp ARE and 6-bp AR ‘half-site' motif 2 matrices in AR-bound sequences. (E) Sequences used in oligonucleotide pull-down assays, corresponding to wild-type (WT) and scrambled (MUT) sequences from the KLK2 promoter 15-bp ARE and UNQ9419 promoter 6-bp AR ‘half-site'. (F) AR western blot of oligonucleotide pull-down fractions, as indicated. Low-molecular-weight region of a parallel Coomassie blue-stained gel shows equal loading of cell lysates. (G,H) UCSC Genome Browser tracks showing 15-bp ARE and 6-bp AR ‘half-site' occurrence in KLK2 and UNQ9419 promoters. (I,K) AR ChIP and qPCR for KLK2 (I) and UNQ9419 (K) promoters from vehicle (+Etok)-and androgen (R1881)-treated experiments. (J,L) Cells were maintained in ethanol (+E) or treated on a time course with androgen (+R; R1881) for 18 h. Material was harvested at 1, 4 and 18 h during the time course and subjected to qrtPCR for KLK (J) and UNQ9419(L), respectively. AR, androgen receptor; ARE, androgen-response element; ChIP, chromatin immunoprecipitation; ChIP-chip, ChIP with on-array detection; Chr, chromosome; KLK2, human glandular kellikrein; qrtPCR, quantitative real-time PCR.
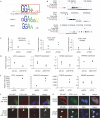
Figure 4
ETS1 and androgen receptor interaction. (A) Sequence logos for ETS1 and AR ChIP-chip-enriched motifs 9 and 1. (B) UCSC Genome Browser tracks showing 15-bp ARE, 6-bp AR ‘half-site' and ETS1 motif occurrence in UNQ9419, CCNG2 and PRAME promoters. Locations of AR ChIP-chip peaks are indicated; darker boxes represent higher signal. (C) ETS1 ChIP and qPCR for UNQ9419, CCNG2 and PRAME promoters from vehicle (+EtOH)- and androgen (R1881)-treated experiments. (D,E) Quantitative real-time PCR of ETS1, UNQ9419, CCNG2 and PRAME 48 h after transfection with an ETS1 RNAi construct (pSR-ETS1) or a scrambled control (pS-Scr) (D) and ETS1 expression construct (+ETS1) or an empty vector control (pSG5) (E). qPCR data were quantified using the standard curve method, values relative to control sample and averages of replicate experiments. (F,G) Confocal microscopy images of LNCaP cells treated with androgen (+R1881) or vehicle (+EtOH) for 1 h. Cells were transfected with either AR–GFP expression construct (F) or pSG5 empty vector (G) and stained for AR (N20) and ETS1 (C20 and 1G11), as indicated. AR, androgen receptor; ARE, androgen-receptor elements; ChIP, chromatin immunoprecipitation; ChIP-chip, ChIP with on-assay detection; DAPI, 4′, 6′-diamidino-2-phenylindole; ETS, avian erythroblastosis virus E26 homologue; GFP, green fluorescent protein; qPCR, quantitative PCR; RNAi, RNA interference.
Similar articles
- DNA specificity determinants associate with distinct transcription factor functions.
Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. Hollenhorst PC, et al. PLoS Genet. 2009 Dec;5(12):e1000778. doi: 10.1371/journal.pgen.1000778. Epub 2009 Dec 18. PLoS Genet. 2009. PMID: 20019798 Free PMC article. - Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome-wide androgen receptor signaling in prostate cancer cells.
Takayama K, Tsutsumi S, Katayama S, Okayama T, Horie-Inoue K, Ikeda K, Urano T, Kawazu C, Hasegawa A, Ikeo K, Gojyobori T, Ouchi Y, Hayashizaki Y, Aburatani H, Inoue S. Takayama K, et al. Oncogene. 2011 Feb 3;30(5):619-30. doi: 10.1038/onc.2010.436. Epub 2010 Oct 4. Oncogene. 2011. PMID: 20890304 - Genome-wide analysis of androgen receptor targets reveals COUP-TF1 as a novel player in human prostate cancer.
Perets R, Kaplan T, Stein I, Hidas G, Tayeb S, Avraham E, Ben-Neriah Y, Simon I, Pikarsky E. Perets R, et al. PLoS One. 2012;7(10):e46467. doi: 10.1371/journal.pone.0046467. Epub 2012 Oct 4. PLoS One. 2012. PMID: 23056316 Free PMC article. - Research Resource: The androgen receptor modulates expression of genes with critical roles in muscle development and function.
Wyce A, Bai Y, Nagpal S, Thompson CC. Wyce A, et al. Mol Endocrinol. 2010 Aug;24(8):1665-74. doi: 10.1210/me.2010-0138. Epub 2010 Jul 7. Mol Endocrinol. 2010. PMID: 20610535 Free PMC article. - Chromatin binding by the androgen receptor in prostate cancer.
Itkonen H, Mills IG. Itkonen H, et al. Mol Cell Endocrinol. 2012 Sep 5;360(1-2):44-51. doi: 10.1016/j.mce.2011.09.037. Epub 2011 Oct 2. Mol Cell Endocrinol. 2012. PMID: 21989426 Review.
Cited by
- Androgen receptor drives transcription of rat PACAP in gonadotrope cells.
Grafer CM, Halvorson LM. Grafer CM, et al. Mol Endocrinol. 2013 Aug;27(8):1343-56. doi: 10.1210/me.2012-1378. Epub 2013 Jun 24. Mol Endocrinol. 2013. PMID: 23798575 Free PMC article. - Allosteric interactions prime androgen receptor dimerization and activation.
Wasmuth EV, Broeck AV, LaClair JR, Hoover EA, Lawrence KE, Paknejad N, Pappas K, Matthies D, Wang B, Feng W, Watson PA, Zinder JC, Karthaus WR, de la Cruz MJ, Hite RK, Manova-Todorova K, Yu Z, Weintraub ST, Klinge S, Sawyers CL. Wasmuth EV, et al. Mol Cell. 2022 Jun 2;82(11):2021-2031.e5. doi: 10.1016/j.molcel.2022.03.035. Epub 2022 Apr 20. Mol Cell. 2022. PMID: 35447082 Free PMC article. - The rules of DNA recognition by the androgen receptor.
Denayer S, Helsen C, Thorrez L, Haelens A, Claessens F. Denayer S, et al. Mol Endocrinol. 2010 May;24(5):898-913. doi: 10.1210/me.2009-0310. Epub 2010 Mar 19. Mol Endocrinol. 2010. PMID: 20304998 Free PMC article. - Coordinate transcriptional regulation by ERG and androgen receptor in fusion-positive prostate cancers.
Chen Y, Sawyers CL. Chen Y, et al. Cancer Cell. 2010 May 18;17(5):415-6. doi: 10.1016/j.ccr.2010.04.022. Cancer Cell. 2010. PMID: 20478521 Free PMC article. - Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer.
Attard G, Clark J, Ambroisine L, Mills IG, Fisher G, Flohr P, Reid A, Edwards S, Kovacs G, Berney D, Foster C, Massie CE, Fletcher A, De Bono JS, Scardino P, Cuzick J, Cooper CS; Transatlantic Prostate Group. Attard G, et al. Br J Cancer. 2008 Jul 22;99(2):314-20. doi: 10.1038/sj.bjc.6604472. Epub 2008 Jul 1. Br J Cancer. 2008. PMID: 18594527 Free PMC article.
References
- Alipov G, Nakayama T, Ito M, Kawai K, Naito S, Nakashima M, Niino D, Sekine I (2005) Overexpression of Ets-1 proto-oncogene in latent and clinical prostatic carcinomas. Histopathology 46: 202–208 - PubMed
- Baillat D, Begue A, Stehelin D, Aumercier M (2002) ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J Biol Chem 277: 29386–29398 - PubMed
- Boyle P, Ferlay J (2005) Cancer incidence and mortality in Europe, 2004. Ann Oncol 16: 481–488 - PubMed
- Carroll JS et al. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 - PubMed
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous