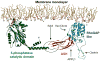A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway - PubMed (original) (raw)
A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway
Kai S Erdmann et al. Dev Cell. 2007 Sep.
Abstract
Mutations in the inositol 5-phosphatase OCRL are responsible for Lowe syndrome, whose manifestations include mental retardation and renal Fanconi syndrome. OCRL has been implicated in membrane trafficking, but disease mechanisms remain unclear. We show that OCRL visits late-stage, endocytic clathrin-coated pits and binds the Rab5 effector APPL1 on peripheral early endosomes. The interaction with APPL1, which is mediated by the ASH-RhoGAP-like domains of OCRL and is abolished by disease mutations, provides a link to protein networks implicated in the reabsorptive function of the kidney and in the trafficking and signaling of growth factor receptors in the brain. Crystallographic studies reveal a role of the ASH-RhoGAP-like domains in positioning the phosphatase domain at the membrane interface and a clathrin box protruding from the RhoGAP-like domain. Our results support a role of OCRL in the early endocytic pathway, consistent with the predominant localization of its preferred substrates, PI(4,5)P(2) and PI(3,4,5)P(3), at the cell surface.
Figures
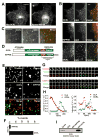
Figure 1. OCRL and the early endocytic pathway
A: Immunofluorescence of the intracellular distribution of Myc-tagged OCRL in Cos-7 cells and comparison with the distribution of co-transfected Xpress-tagged INPP5B. B: RFP-OCRL (and RFP-INPP5B) colocalizes with EGFP-rab5 positive endosomes but not with EGFP-Rab9-positive organelles. C: EGFP-OCRL positive spots are directly adjacent to endogenous retromer (SNX1) immunoractivity suggesting budding of retromer positive membrane from OCRL positive endosomes. D: Modular structure of OCRL and INPP5B. E: TIRF microscopy of Cos-7 cells transfected with EGFP-clathrin light chain together with RFP-OCRL (left column) or RFP-INPP5B (right column). OCRL, but not INPP5B, colocalizes with a subset of clathrin spots in these single short exposure (200 msec) frames. F: Percentage of clathrin coated pits positive for INPP5B or for OCRL during their lifetime. G and H: Sequential TIRF microscopy images (8 sec intervals)(G) and time course of fluorescence intensity (H) for two clathrin-coated pits from cells co-transfected with RFP-clathrin and EGFP-OCRL. Note: In H, fluorescence is measured at a specific position so that both loss of fluorescence and lateral movement of the fluorescent spot away from such position result in a loss of signal intensity. I: Interaction of clathrin heavy chain with OCRL but not with INPP5B as revealed by western blotting of material affinity-purified by GST-OCRL or GST-INPP5B from a rat brain lysate (Bars = A, 10 μm; B, 2.5 μm; C, 5 μm; E, 5 μm; F, 1 μm).
Figure 2. The Rab5 effector protein APPL1 interacts, and colocalizes with, OCRL and INPP5B
A: A 90 kd protein binds OCRL and INPP5B in pulldown experiments. GST and GST fusion proteins of the COOH-terminal regions of OCRL and INPP5B were incubated with rat brain lysate and bound proteins were analyzed by SDS-PAGE followed by Coomassie blue staining. B: Anti-APPL1 western blotting of the samples shown in A as well as bound proteins from pulldowns in which GST-fusions of full length INPP5B and OCRL were used as baits. C: The interaction between OCRL/INPP5B and APPL1 is direct. His-tagged recombinant APPL1 was incubated with the indicated GST-fusion proteins and bound proteins were analyzed by western-blotting. D: OCRL and INPP5B interact with APPL1 but not with APPL2 as shown by GST-pulldowns from transfected cells followed by western blotting. E: Coprecipitation of OCRL with APPL1 from extracts of transfected Cos-7 cells. Western blotting of the starting lysate and of the immunoprecipitates are shown. F: Epifluorescence images demonstrating that APPL1 colocalizes with OCRL and INPP5B on endosomes in transfected Cos-7 cells. G: H and I: TIRF microscopy sequential images (12 sec intervals) of spots positive for RFP-clathrin, EGF-OCRL and EGFP- or RFP-APPL1 at the cell surface (Bars = F, 2.5 μm; G, H and I = 1μm).
Figure 3. OCRL and APPL1 are components of a protein network implicated in receptor trafficking and signalling
A: Schematic drawing indicating direct and indirect interactions of OCRL described in this study or reported in the literature. The PDZ domain of GIPC binds the COOH-terminus of both the TrkA receptor and megalin. The PTB domain of APPL1 also binds the cytoplasmic domain of TrkA. A direct interaction of this PTB domain with megalin remains hypothetical. B: GST pulldowns of a rat brain lysate on GST or GST fusions of the COOH-terminal region of OCRL (top) or of the cytoplasmic domain of megalin (bottom). The western blots shown demonstrate presence of both APPL1 and GIPC in the materials bound both baits. C: Protein network involving OCRL and APPL1 and potentially relevant to the kidney and brain defects observed in Lowe syndrome. Loss of function mutations of proteins indicated in red lead to renal proximal tubule reabsorption defects either in human (OCRL) or in mouse (Megalin, GIPC and Dab2)(see text). D: Presence of APPL1 in both rat brain and kidney lysates (western blot). E: Immunofluorescence of cross-sectioned (cryostat sections) mouse kidney proximal tubules showing enrichment of APPL1 at the apical region of the epithelium, where it colocalizes with GIPC, myosin VI, and megalin, and also, partially with γ-adaptin. (Bar = 10 μm)
Figure 4. Mapping of the binding interfaces between APPL1 and OCRL
A: Modular structure and interactions of APPL1. The figure also shows an alignment of the 11-mer peptide of human APPL1 with the corresponding sequence of APPL from other species, demonstrating its evolutionary conservation. The sequence is not conserved in human APPL2. Potential phosphorylation sites within the 11-mer peptide are indicated by asterisks. B: GST pulldowns from extracts of Cos-7 expressing either Myc-OCRL or Xpress-INPP5B define an 11 amino acid (residues 403–413) stretch of human APPL1 as the minimal region necessary and sufficient for binding. Bound proteins were identified by western blotting. Numbers indicate the amino acid boundaries of the APPL1 fragment fused to GST. The F404A mutation abolished binding. C: ITC analysis of the binding of the wildtype APPL1 11-mer peptide and of the F404A-mutant peptide. Raw data are shown in the upper panels, and plots of the total heat released as a function of the molar ratio of each ligand are shown in the bottom panels. D: GST-fusions of the wildtype or mutant (S403D and S410D) minimal binding region (11-mer peptide) were incubated with the catalytic subunit of PKA or with type II Ca2+-calmodulin dependent kinase (CamKII) in the presence of γ-32P-ATP. Proteins were separated by SDS-PAGE and the corresponding autoradiography is shown. E: Co-immunoprecipitation of full length wildtype and mutant (S410D) APPL1 with OCRL. F: PKA overexpression impairs binding of APPL1 to OCRL in vivo. For E and F Cos-7 cells were transfected with the indicated expression constructs. Protein complexes were immunoprecipitated using anti-GFP antibody and the starting lysates and the immunoprecipitates were analyzed by western blotting.
Figure 5. Structure of the COOH-terminal region of OCRL
A: Ribbon diagram with the ASH domain (red) and the RhoGAP-like domain (blue). Residues of the clathrin box motif are in sticks. B: Stereo view of the Cα backbone traces. Trace color ramped from the NH2 terminus (red) to the COOH-terminus (blue). Every 20 residues are labeled and indicated with small filled circles. C: (Left) RhoGAP like domain of OCRL (blue) superimposed on the RhoGAP domain of P50RhoGAP (PDB ID: 1AM4). Bound Cdc42 is shown in gold. (Right) Biochemical interaction of the RhoGAP-like domain of OCRL and INPP5B with Rac and Cdc42. GST pulldowns from lysates of Cos-7 cells expressing tagged versions of Rac, Rho or Cdc42 with GST-fusions of the RhoGAP-like domain of OCRL or INPP5B. Bound proteins were analyzed by western-blotting. D: Alignment of the ASH domain of OCRL and INPP5B with other members of the MSP/VAP domain protein family. The E585 deletion mutation found in a patient is marked by a black triangle. Secondary structure elements are drawn above the alignment. Entrez database accession numbers are as follows: OCRL_Hs, GI: 57209431; Inpp5_Hs, GI: 59803021; OCRL_Dr, GI: 68374521; Inpp5_Dm, GI: 54642833; Inpp5_Ce, GI: 17505597; Inpp5_Dd, GI: 66828629; MSP_Ce, GI: 21730216; Vap-A_Rn, GI: 73535850. (Hs, Homo sapiens; Dr, Danio rerio; DM, Drosophila melanogaster; CE, Caenorhabditis elegans; Dd_, Dictyostelium discoideum;_ Rn, Rattus norvegicus).
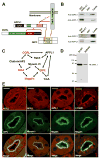
Figure 6. OCRL point mutations of Lowe syndrome patients abolish the interaction with APPL1
A: Close view of the local structural environment of three single amino acid patient mutations in the COOH-terminal regio n of OCRL. B: GST-pulldowns from extracts of rat brain (APPL1, clathrin) or transfected Cos-7 cells (Myc-RacV12) on wildtype and mutant fusion proteins, demonstrating that each of the three mutations abolish binding to APPL1 but not to clathrin or Rac (Myc-RacV12). Bound proteins were detected by western blotting. C: Two colour fluorescence of cells expressing RFP-APPL1 and EGFP-OCRL (wildtype or mutant). Wildtype OCRL is localized on spots throughout the cell. The more peripheral spots (arrowheads), but not all spots (arrows), colocalize with APPL1. Each of three patient mutations abolishes the colocalization with APPL1, although APPL1 positive spots at the periphery persist. Note that at least for the Δ585 mutation, OCRL still has a punctate subcellular distribution.
Figure 7. Structural model of the interaction of OCRL and INPP5B with the membrane
The ASH-RhoGAP-like domains of OCRL were docked to the structure of the inositol 5-phosphastase domain from S. Pombe determined by Hurley and coworkers (Tsujishita et al., 2001). The NH2-terminal domain of OCRL, whose structure remains unknown, is not included. OCRL and INPP5B form an arc that is anchored to the membrane via the catalytic site of the inositol 5-phosphatase domain (shown here in a complex with the head group of a phosphoinositide) (PDB ID: 1I9Z) and the COOH-terminal region of the RhoGAP-like domain, which in INPP5B is farnesylated. Three mutations which abolish Rab5 binding are located within a small region (solid oval line), whose distance from the bilayer can accommodate a small GTPase such as Rab5. Rac or Cdc42 could also be accommodated next to the RhoGAP-like domain. The precise binding interface for APPL1 (dashed oval line) remains to be defined. The clathrin binding site protrudes from the structure.
Similar articles
- All known patient mutations in the ASH-RhoGAP domains of OCRL affect targeting and APPL1 binding.
McCrea HJ, Paradise S, Tomasini L, Addis M, Melis MA, De Matteis MA, De Camilli P. McCrea HJ, et al. Biochem Biophys Res Commun. 2008 May 2;369(2):493-9. doi: 10.1016/j.bbrc.2008.02.067. Epub 2008 Feb 26. Biochem Biophys Res Commun. 2008. PMID: 18307981 Free PMC article. - Two closely related endocytic proteins that share a common OCRL-binding motif with APPL1.
Swan LE, Tomasini L, Pirruccello M, Lunardi J, De Camilli P. Swan LE, et al. Proc Natl Acad Sci U S A. 2010 Feb 23;107(8):3511-6. doi: 10.1073/pnas.0914658107. Epub 2010 Feb 2. Proc Natl Acad Sci U S A. 2010. PMID: 20133602 Free PMC article. - A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism.
Mao Y, Balkin DM, Zoncu R, Erdmann KS, Tomasini L, Hu F, Jin MM, Hodsdon ME, De Camilli P. Mao Y, et al. EMBO J. 2009 Jul 8;28(13):1831-42. doi: 10.1038/emboj.2009.155. Epub 2009 Jun 18. EMBO J. 2009. PMID: 19536138 Free PMC article. - The role of the Lowe syndrome protein OCRL in the endocytic pathway.
Sharma S, Skowronek A, Erdmann KS. Sharma S, et al. Biol Chem. 2015 Dec;396(12):1293-300. doi: 10.1515/hsz-2015-0180. Biol Chem. 2015. PMID: 26351914 Review. - The 5-phosphatase OCRL in Lowe syndrome and Dent disease 2.
De Matteis MA, Staiano L, Emma F, Devuyst O. De Matteis MA, et al. Nat Rev Nephrol. 2017 Aug;13(8):455-470. doi: 10.1038/nrneph.2017.83. Epub 2017 Jul 3. Nat Rev Nephrol. 2017. PMID: 28669993 Review.
Cited by
- Impaired neural development in a zebrafish model for Lowe syndrome.
Ramirez IB, Pietka G, Jones DR, Divecha N, Alia A, Baraban SC, Hurlstone AF, Lowe M. Ramirez IB, et al. Hum Mol Genet. 2012 Apr 15;21(8):1744-59. doi: 10.1093/hmg/ddr608. Epub 2011 Dec 30. Hum Mol Genet. 2012. PMID: 22210625 Free PMC article. - Species-specific difference in expression and splice-site choice in Inpp5b, an inositol polyphosphate 5-phosphatase paralogous to the enzyme deficient in Lowe Syndrome.
Bothwell SP, Farber LW, Hoagland A, Nussbaum RL. Bothwell SP, et al. Mamm Genome. 2010 Oct;21(9-10):458-66. doi: 10.1007/s00335-010-9281-7. Epub 2010 Sep 26. Mamm Genome. 2010. PMID: 20872266 Free PMC article. - Dent's disease: clinical features and molecular basis.
Claverie-Martín F, Ramos-Trujillo E, García-Nieto V. Claverie-Martín F, et al. Pediatr Nephrol. 2011 May;26(5):693-704. doi: 10.1007/s00467-010-1657-0. Epub 2010 Oct 10. Pediatr Nephrol. 2011. PMID: 20936522 - PTEN reduces endosomal PtdIns(4,5)P2 in a phosphatase-independent manner via a PLC pathway.
Mondin VE, Ben El Kadhi K, Cauvin C, Jackson-Crawford A, Bélanger E, Decelle B, Salomon R, Lowe M, Echard A, Carréno S. Mondin VE, et al. J Cell Biol. 2019 Jul 1;218(7):2198-2214. doi: 10.1083/jcb.201805155. Epub 2019 May 22. J Cell Biol. 2019. PMID: 31118240 Free PMC article. - Class I and class III phosphoinositide 3-kinases are required for actin polymerization that propels phagosomes.
Bohdanowicz M, Cosío G, Backer JM, Grinstein S. Bohdanowicz M, et al. J Cell Biol. 2010 Nov 29;191(5):999-1012. doi: 10.1083/jcb.201004005. J Cell Biol. 2010. PMID: 21115805 Free PMC article.
References
- Astle MV, Seaton G, Davies EM, Fedele CG, Rahman P, Arsala L, Mitchell CA. Regulation of phosphoinositide signaling by the inositol polyphosphate 5-phosphatases. IUBMB Life. 2006;58:451–456. - PubMed
- Attree O, Olivos IM, Okabe I, Bailey LC, Nelson DL, Lewis RA, McInnes RR, Nussbaum RL. The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–242. - PubMed
- Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. - PubMed
- Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. - PubMed
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5T32GM07205/GM/NIGMS NIH HHS/United States
- P01 CA046128/CA/NCI NIH HHS/United States
- P30 DA018343/DA/NIDA NIH HHS/United States
- DK45735/DK/NIDDK NIH HHS/United States
- CA46128/CA/NCI NIH HHS/United States
- P30 DK045735/DK/NIDDK NIH HHS/United States
- DA018343/DA/NIDA NIH HHS/United States
- NS36251/NS/NINDS NIH HHS/United States
- R37 NS036251/NS/NINDS NIH HHS/United States
- R01 NS036251/NS/NINDS NIH HHS/United States
- T32 GM007205/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous