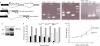T-cell factor 4 (Tcf7l2) maintains proliferative compartments in zebrafish intestine - PubMed (original) (raw)
T-cell factor 4 (Tcf7l2) maintains proliferative compartments in zebrafish intestine
Vanesa Muncan et al. EMBO Rep. 2007 Oct.
Abstract
Previous studies have shown that Wnt signals, relayed through beta-catenin and T-cell factor 4 (Tcf4), are essential for the induction and maintenance of crypts in mice. We have now generated a tcf4 (tcf7l2) mutant zebrafish by reverse genetics. We first observe a phenotypic defect at 4 weeks post-fertilization (wpf), leading to death at about 6 wpf. The phenotype comprises a loss of proliferation at the base of the intestinal folds of the middle and distal parts of the intestine. The proximal intestine represents an independent compartment, as it expresses sox2 in the epithelium and barx1 in the surrounding mesenchyme, which are early stomach markers in higher vertebrates. Zebrafish are functionally stomach-less, but the proximal intestine might share its ontogeny with the mammalian stomach. Rare adult homozygous tcf4(-/-) 'escapers' show proliferation defects in the gut epithelium, but have no other obvious abnormalities. This study underscores the involvement of Tcf4 in maintaining proliferative self-renewal in the intestine throughout life.
Figures
Figure 1
Analysis of splice/donor mutation in the zebrafish tcf4 gene, showing poor survival and slower growth rate in _tcf4_exI/exI fish. (A) Diagram of the _tcf4_exI/exI allele. The arrowhead points to nucleic acid substitution in the donor splice site of the first intron. The position of a stop codon within the first intron is underlined. Positions and PCR products of primer sets F1/R1 and F2/R2 are indicated. Note that primer R2 overlaps the exon I/exon II boundary. (B) RT–PCR preformed on complementary DNA samples from 7 dpf embryos with the primer set F1/R1. Heterozygous tcf4 cDNA (_tcf4_exI/wt) yields two bands (lane 2). The upper mutant band contains the retained intron. _tcf4_wt/wt (lane 3); _tcf4_exI/exI (lane 1). (C) RT–PCR of adult homozygous ‘escapers' with the primer set F1/R1 (lane 2). In mutant ‘escapers', only the upper band in which the intron is retained is present. Heterozygous (lane 1); wild-type (wt; lane 3). (D) RT–PCR using the primer combination F2/R2. No wt band can be amplified in either tcf4exI/exI embryos (lane 1) or adult mutant escapers (lane 2). The same primer set detects only the wt band in heterozygous fish (lane 3). cDNA from wt fish (lane 4). (E) Western blot analysis of Tcf4 protein on 7 dpf _tcf4_exI/exI (right lane) or _tcf4_wt/wt (left lane) larvae. (F) Survival of the _tcf4_exI/exI genotype (black bars) in time (weeks). The _tcf4_exI/exI genotype is significantly lost first at 6 wpf. White bars represent the genotype frequency of wild types (_tcf4_wt/wt) and grey bars that of heterozygotes (_tcf4_exI/wt). (G) Body length of _tcf4_wt/wt and _tcf4_exI/exI fish monitored weekly. Mutant fish grow slower after 3 wpf. Data are percentages (F) or mean±s.d. (G). See Methods. All reference to phenotypes was confirmed by genotyping (see Methods). dpf, days post-fertilization; RT–PCR, reverse transcription–PCR; Tcf4, T-cell factor 4; wpf, weeks post-fertilization.
Figure 2
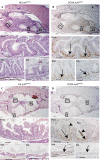
The absence of cycling cells in _tcf4_exI/exI at 6 weeks post-fertilization. (A) Haematoxylin and eosin (HE) staining of _tcf4_wt/wt zebrafish intestine with the proximal (Prox), middle (Mid) and distal (Dis) parts magnified respectively in lower panels. (B) PCNA staining on consecutive paraffin sections of wild-type (wt) fish. The arrows in the magnified lower panels indicate PCNA+ cells in the interfold pockets. (C) HE staining of _tcf4_exI/exI intestine. (D) PCNA staining on consecutive sections depicting absent proliferation in the middle and distal intestinal portions. Note that proliferation in the proximal intestine is maintained (arrows and arrowheads in the magnified proximal intestine in (D)). PCNA, proliferating cell nuclear antigen; wpf, weeks post-fertilization. Scale bars: top panels (A–D), 250 μm; middle and bottom panels (A–D), 50 μm.
Figure 3
Absence of BrdU-labelled cells in the middle and distal intestine in _tcf4_exI/exI fish at 5 weeks post-fertilization. (A–F) Anti-BrdU immunohistochemistry of _tcf4_wt/wt at 2 (A), 3 (C) and 5 wpf (E) and _tcf4_exI/exI at 2 (B), 3 (C) and 5 wpf (F). BrdU-labelled cells after 3 h pulse are observed throughout the intestinal folds at 2 and 3 wpf in mutant and wild-type (wt) fish (black arrows in (A–D)). At 5 wpf, BrdU-labelled cells occur between folds (black arrows in (E)). In the mutant fish, proliferation in the proximal (Prox) intestine is not restricted to the interfold pockets (black arrows in the middle panel of (F)). No BrdU-labelled cells are observed in the middle (Mid) and distal (Dis) intestine in mutant fish (F, bottom panel). (G) BrdU cells are scored as a percentage of total cell number per analysed segment. Error bars are mean±s.d. The asterisk indicates statistically significant difference (Student's _t_-test, _P_=0.025). At least three fish were analysed per time point per genotype. BrdU, 5-bromo-2-deoxyuridine; wpf, weeks post-fertilization.
Figure 4
The proximal part of zebrafish intestine is molecularly marked as stomach. (A) A wild-type (wt) embryo at 96 hpf with oesophagus (Eso) and intestine (Int) boundaries corresponding to position of somites II and III. The inset indicates the difference in epithelial morphology between oesophagus and intestine, corresponding to somites II and III. (B) WISH for sox2 messenger RNA, expressed in the most proximal region (arrows), corresponding to somite III. (C,E) gata5 and gata6 expression starts from somite IV. (D) WISH for barx1 mRNA, depicting specific mesenchymal expression around the proximal intestinal segment, corresponding to somite III. (F) Scheme of expression domains correlated to somites at 96 hpf. hpf, hours post-fertilization; WISH, whole-mount in situ hybridization; L, liver; PhA, pharyngeal arches; SB, swimming bladder.
Figure 5
Loss of cdx4 induces ectopic expression of proximal intestinal markers in the middle intestine. WISH for sox2 and barx1 in wild type (wt; A,C) and kgg tv205 mutants (B,D) is shown. The arrows point to sox2 and barx1 expression. WISH, whole-mount in situ hybridization.
Figure 6
Gut phenotype of homozygous Tcf4 mutant ‘escaper' fish. (A) Escaper compared with sibling is reduced in size. (B) Western blot analysis of Tcf4 protein from brain and fin tissues showing the absence of protein in escapers. (C) PCNA IHC in wild type (wt). The lower panel is a magnification of the boxed area; the arrows point to PCNA+ cells at the base of the folds. (D) PCNA IHC on Tcf4 escaper fish showing large areas of intestinal epithelium with loss of proliferation. The three lower panels are magnifications of the boxed areas. The arrows point to severely affected areas or to rare PCNA+ patches unequally distributed throughout the epithelial folds. The dashed line indicates the flat epithelial layer in this fish. IHC, immunohistochemistry; PCNA, proliferating cell nuclear antigen; Tcf-4, T-cell factor.
References
- Beck F, Chawengsaksophak K, Luckett J, Giblett S, Tucci J, Brown J, Poulsom R, Jeffery R, Wright NA (2003) A study of regional gut endoderm potency by analysis of Cdx2 mutant chimaeric mice. Dev Biol 255: 399–406 - PubMed
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W (1996) Functional interaction of β-catenin with the transcription factor LEF-1. Nature 382: 638–642 - PubMed
- Cho EA, Dressler GR (1998) TCF-4 binds β-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech Dev 77: 9–18 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases