Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis - PubMed (original) (raw)
Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis
David E Greenberg et al. J Bacteriol. 2007 Dec.
Abstract
Chronic granulomatous disease (CGD) is an inherited immune deficiency characterized by increased susceptibility to infection with Staphylococcus, certain gram-negative bacteria, and fungi. Granulibacter bethesdensis, a newly described genus and species within the family Acetobacteraceae, was recently isolated from four CGD patients residing in geographically distinct locales who presented with fever and lymphadenitis. We sequenced the genome of the reference strain of Granulibacter bethesdensis, which was isolated from lymph nodes of the original patient. The genome contains 2,708,355 base pairs in a single circular chromosome, in which 2,437 putative open reading frames (ORFs) were identified, 1,470 of which share sequence similarity with ORFs in the nonpathogenic but related Gluconobacter oxydans genome. Included in the 967 ORFs that are unique to G. bethesdensis are ORFs potentially important for virulence, adherence, DNA uptake, and methanol utilization. GC% values and best BLAST analysis suggested that some of these unique ORFs were recently acquired. Comparison of G. bethesdensis to other known CGD pathogens demonstrated conservation of some putative virulence factors, suggesting possible common mechanisms involved in pathogenesis in CGD. Genotyping of the four patient isolates by use of a custom microarray demonstrated genome-wide variations in regions encoding DNA uptake systems and transcriptional regulators and in hypothetical ORFs. G. bethesdensis is a genetically diverse emerging human pathogen that may have recently acquired virulence factors new to this family of organisms.
Figures
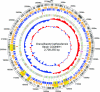
FIG. 1.
Genome circle of Granulibacter bethesdensis. The genome is 2,708,355 base pairs long, and the rings, starting inside and moving out, show the following: GC content with sliding 20-kb window; GC skew; ORFs colored by metabolic category (virulence = turquoise, cell wall metabolism = light yellow, stress = light orange, phage = orange, signal transduction = purple, cellular processing = yellow, secondary metabolism = blue, membrane transport = cyan, secretion = teal, motility and chemotaxis = pink, information processing = sage, phosphorus metabolism = olive, nitrogen metabolism = tangerine, coenzyme and cofactor metabolism = saffron, sulfur metabolism = light green, bioenergetics = lime, one-carbon metabolism = beige, aromatic compound degradation = light purple, nucleotide metabolism = light blue, lipid metabolism = lemon, carbohydrate metabolism = rust, amino acid metabolism = peach); RNAs (tRNAs are shown in green, and rRNAs are shown in red); ORFs colored and positioned by orientation (orange, positive strand; and blue, negative strand); yellow shaded areas representing regions undergoing DNA changes across the four isolates, based on chip hybridization data; ORFs present in G. bethesdensis that are unique compared to G. oxydans (black bars); and numeric markers.
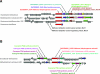
FIG. 2.
Comparative genomic context of Granulibacter bethesdensis methanol dehydrogenase genes. A comparative region analysis of two methanol dehydrogenase subunit 1 ORFs in G. bethesdensis compared to other best-hit genes and genomes is shown. (A) GbCGDNIH1_0344 and surrounding genes and their relationships to those in other methylotrophic organisms. (B) GbCGDNIH1_1922 and surrounding genes needed for methanol utilization and their relationship to the methylotrophic bacterium Methylobacterium extorquens AM1.
FIG. 3.
Quantitative PCR analysis of six different Granulibacter bethesdensis gene mRNAs at 35°C in the stationary phase of growth. Bacteria were cultured in YPG broth at 35°C to stationary phase. Multiplex quantitative PCR was performed to measure the transcripts from six different G. bethesdensis genes, and their signals were normalized to the mRNA signal from the transcript for the protein translation elongation factor TU (tufA) gene. The y axis shows relative mRNA levels for each of the six genes compared to tufA, and the x axis shows the six individual genes tested.
FIG. 4.
Ultrastructural evidence of extracellular polysaccharide capsule and conjugative pilus-like structures. (A and B) Transmission electron micrographs of G. bethesdensis NIH1.1 prepared with (A) and without (B) ruthenium red for preserving and contrasting extracellular polysaccharides. The arrow in panel A points out the capsular material on the surface. (C and D) Transmission (C) and scanning (D) electron micrographs of structures resembling conjugative pili. Bars, 100 nm.
FIG. 5.
Dendrogram of DNA hybridization of seven isolates with NIH1.1. A DNA-DNA hybridization microarray-based investigation of gene distributions among G. bethesdensis isolates was performed. The absence of genes is shown in blue, and the presence of genes is shown in yellow and red. A dendrogram is shown with the strain names listed. The analysis was performed with GeneSpring software.
Similar articles
- Recurrent Granulibacter bethesdensis infections and chronic granulomatous disease.
Greenberg DE, Shoffner AR, Zelazny AM, Fenster ME, Zarember KA, Stock F, Ding L, Marshall-Batty KR, Wasserman RL, Welch DF, Kanakabandi K, Sturdevant DE, Virtaneva K, Porcella SF, Murray PR, Malech HL, Holland SM. Greenberg DE, et al. Emerg Infect Dis. 2010 Sep;16(9):1341-8. doi: 10.3201/eid1609.091800. Emerg Infect Dis. 2010. PMID: 20735916 Free PMC article. - Simultaneous Host-Pathogen Transcriptome Analysis during Granulibacter bethesdensis Infection of Neutrophils from Healthy Subjects and Patients with Chronic Granulomatous Disease.
Greenberg DE, Sturdevant DE, Marshall-Batty KR, Chu J, Pettinato AM, Virtaneva K, Lane J, Geller BL, Porcella SF, Gallin JI, Holland SM, Zarember KA. Greenberg DE, et al. Infect Immun. 2015 Nov;83(11):4277-92. doi: 10.1128/IAI.00778-15. Epub 2015 Aug 17. Infect Immun. 2015. PMID: 26283340 Free PMC article. - Serologic reactivity to the emerging pathogen Granulibacter bethesdensis.
Greenberg DE, Shoffner AR, Marshall-Batty KR, Arora K, Zhao M, Martin R, Ding L, Hammer CH, Shaw PA, Kuhns DB, Malech HL, Gallin JI, Zarember KA, Holland SM. Greenberg DE, et al. J Infect Dis. 2012 Sep 15;206(6):943-51. doi: 10.1093/infdis/jis431. Epub 2012 Jul 10. J Infect Dis. 2012. PMID: 22782953 Free PMC article. - Review of virulence factors of enterococcus: an emerging nosocomial pathogen.
Giridhara Upadhyaya PM, Ravikumar KL, Umapathy BL. Giridhara Upadhyaya PM, et al. Indian J Med Microbiol. 2009 Oct-Dec;27(4):301-5. doi: 10.4103/0255-0857.55437. Indian J Med Microbiol. 2009. PMID: 19736397 Review. - Microbial genome sequencing and pathogenesis.
Tang CM, Hood DW, Moxon ER. Tang CM, et al. Curr Opin Microbiol. 1998 Feb;1(1):12-6. doi: 10.1016/s1369-5274(98)80137-9. Curr Opin Microbiol. 1998. PMID: 10066467 Review.
Cited by
- A draft genome sequence and functional screen reveals the repertoire of type III secreted proteins of Pseudomonas syringae pathovar tabaci 11528.
Studholme DJ, Ibanez SG, MacLean D, Dangl JL, Chang JH, Rathjen JP. Studholme DJ, et al. BMC Genomics. 2009 Aug 24;10:395. doi: 10.1186/1471-2164-10-395. BMC Genomics. 2009. PMID: 19703286 Free PMC article. - Granulibacter bethesdensis, a Pathogen from Patients with Chronic Granulomatous Disease, Produces a Penta-Acylated Hypostimulatory Glycero-D-talo-oct-2-ulosonic Acid-Lipid A Glycolipid (Ko-Lipid A).
Muszyński A, Zarember KA, Heiss C, Shiloach J, Berg LJ, Audley J, Kozyr A, Greenberg DE, Holland SM, Malech HL, Azadi P, Carlson RW, Gallin JI. Muszyński A, et al. Int J Mol Sci. 2021 Mar 24;22(7):3303. doi: 10.3390/ijms22073303. Int J Mol Sci. 2021. PMID: 33804872 Free PMC article. - Gluconobacter as well as Asaia species, newly emerging opportunistic human pathogens among acetic acid bacteria.
Alauzet C, Teyssier C, Jumas-Bilak E, Gouby A, Chiron R, Rabaud C, Counil F, Lozniewski A, Marchandin H. Alauzet C, et al. J Clin Microbiol. 2010 Nov;48(11):3935-42. doi: 10.1128/JCM.00767-10. Epub 2010 Sep 8. J Clin Microbiol. 2010. PMID: 20826638 Free PMC article. - The urea carboxylase and allophanate hydrolase activities of urea amidolyase are functionally independent.
Lin Y, Boese CJ, St Maurice M. Lin Y, et al. Protein Sci. 2016 Oct;25(10):1812-24. doi: 10.1002/pro.2990. Epub 2016 Aug 5. Protein Sci. 2016. PMID: 27452902 Free PMC article. - A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti.
Mullins EA, Francois JA, Kappock TJ. Mullins EA, et al. J Bacteriol. 2008 Jul;190(14):4933-40. doi: 10.1128/JB.00405-08. Epub 2008 May 23. J Bacteriol. 2008. PMID: 18502856 Free PMC article.
References
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. - PubMed
- Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous